| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,511,166 – 24,511,266 |
| Length | 100 |
| Max. P | 0.975880 |

| Location | 24,511,166 – 24,511,266 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.58031 |
| G+C content | 0.51945 |
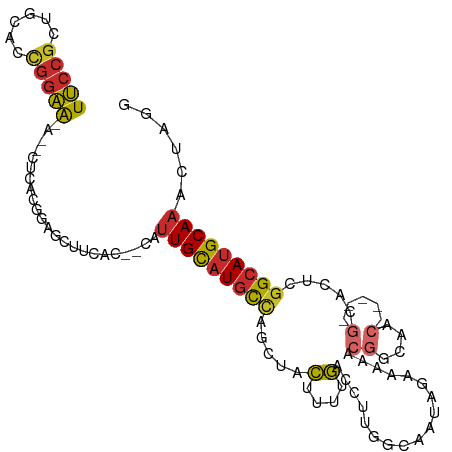
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -13.48 |
| Energy contribution | -13.72 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
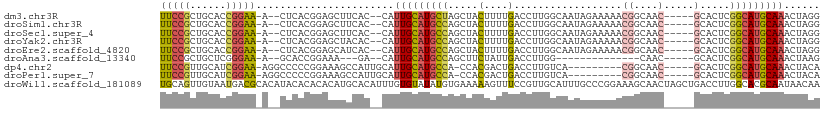
>dm3.chr3R 24511166 100 + 27905053 UUCCGCUGCACCGGAA-A--CUCACGGAGCUUCAC--CAUUGCAUGCUAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC-----GCACUCGGCAUGCAAACUAGG ..(((..((.(((...-.--....))).))..)..--..(((((((((.((((..........))))..........((....)-----).....)))))))))....)) ( -26.30, z-score = -1.40, R) >droSim1.chr3R 24208571 100 + 27517382 UUCCGCUGCACCGGAA-A--CUCACGGAGCUUCAC--CAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC-----GCACUCGGCAUGCAAACUAGG ..(((..((.(((...-.--....))).))..)..--..(((((((((.((((..........))))..........((....)-----).....)))))))))....)) ( -29.00, z-score = -2.14, R) >droSec1.super_4 3393188 100 + 6179234 UUCCGCUGCACCGGAA-A--CUCACGGAGCUUCAC--CAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC-----GCACUCGGCAUGCAAACUAGG ..(((..((.(((...-.--....))).))..)..--..(((((((((.((((..........))))..........((....)-----).....)))))))))....)) ( -29.00, z-score = -2.14, R) >droYak2.chr3R 6853227 100 - 28832112 UUCCGCUGCACCGGAA-A--CUCACGGAGCUACAC--CAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC-----GCACUCGGCAUGCAAACUAGG ..(((..((.(((...-.--....))).))..)..--..(((((((((.((((..........))))..........((....)-----).....)))))))))....)) ( -29.00, z-score = -2.34, R) >droEre2.scaffold_4820 6954083 100 - 10470090 UUCCGCUGCACCGGAA-A--CUCACGGAGCAUCAC--CAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC-----GCACUCGGCAUGCAAACUAGG ..(((.(((.(((...-.--....))).))).)..--..(((((((((.((((..........))))..........((....)-----).....)))))))))....)) ( -30.30, z-score = -2.66, R) >droAna3.scaffold_13340 17317676 83 + 23697760 UUCCGCUGCUCGGGAA-A--GCACCGGAAA---GA--CAUUGCAUGCCAGCUUCUAUUGACCUUGG--------------CAAC-----GCACUCGGCAUGCAAACUAAG (((((.((((......-)--))).))))).---..--..(((((((((....((....))....(.--------------...)-----......)))))))))...... ( -27.00, z-score = -2.31, R) >dp4.chr2 27289668 94 - 30794189 UUCCGUUGCAUCGGAA-AGGCCCCCGGAAAGCCAUUGCAUUGCAUGCCA-CCACGACUGACCUUGUCA---------CGGCAAC-----GCACUCGGCAUGCAAACUACA (((((......)))))-.(((.........)))......(((((((((.-....(((.......))).---------((....)-----).....)))))))))...... ( -30.30, z-score = -1.86, R) >droPer1.super_7 840113 94 + 4445127 UUCCGUUGCAUCGGAA-AGGCCCCCGGAAAGCCAUUGCAUUGCAUGCCA-CCACGACUGACCUUGUCA---------CGGCAAC-----GCACUCGGCAUGCAAACUACA (((((......)))))-.(((.........)))......(((((((((.-....(((.......))).---------((....)-----).....)))))))))...... ( -30.30, z-score = -1.86, R) >droWil1.scaffold_181089 9223691 110 + 12369635 UGCAGUUGUAAUGACGCACAUACACACACAUGCACAUUUGUGUAUAUGUGAAAAAGUUUCCGUUGCAUUUGCCCGGAAAGCAACUAGCUGACCUUGGCACGCAAUAACAA (((((((((.......((((((.(((((.((....)).))))).))))))......((((((..((....)).)))))))))))).(((......)))..)))....... ( -31.90, z-score = -1.57, R) >consensus UUCCGCUGCACCGGAA_A__CUCACGGAGCUUCAC__CAUUGCAUGCCAGCUACUUUUGACCUUGGCAAUAGAAAAACGGCAAC_____GCACUCGGCAUGCAAACUAGG .((((......))))........................(((((((((.....(....)....................((........))....)))))))))...... (-13.48 = -13.72 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:52:01 2011