
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,500,532 – 24,500,629 |
| Length | 97 |
| Max. P | 0.995003 |

| Location | 24,500,532 – 24,500,629 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.55935 |
| G+C content | 0.34526 |
| Mean single sequence MFE | -15.08 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.995003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
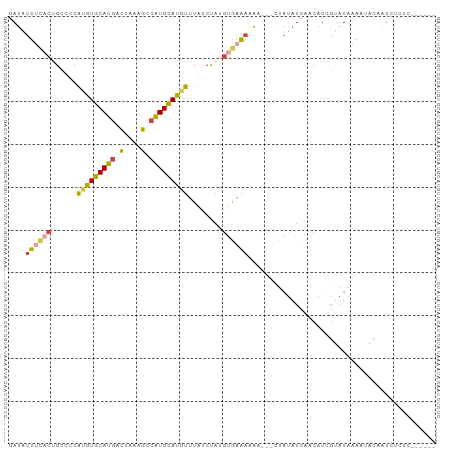
>dm3.chr3R 24500532 97 + 27905053 GAUAUUUCACUGCUCCAUGUGCAUGACUAAAGCCAUGCAUGUUUAUCUAUGUGAACAAA---CUAUAUCAACAUCGUAUAAAAAACAAUCUCUCUCUUCC ((((((((((......(((((((((........)))))))))........)))))....---..)))))............................... ( -15.14, z-score = -2.18, R) >droSim1.chr3R 24197959 97 + 27517382 GAUAUUUCACUGCUCCAUGUGCAUGACUAAAGCCAUGCAUGUUUAUCUAUGUGAAAAAA---CUAUAUCAACAUCGUAUAAAAUACAAUCACUCUCUUCC (((.((((((......(((((((((........)))))))))........))))))...---.............((((...)))).))).......... ( -15.94, z-score = -2.30, R) >droYak2.chr3R 6842571 91 - 28832112 GAUAUUUCACUGCCUCAUGUGCAUGACUAAAGCCAUGCAUGUUUAGCCAUGUGAAAAAA---CUAUAUCAACAUCGUAUAAAAUACAAUCUCUC------ ((((((((((.((...(((((((((........)))))))))...))...)))))....---..)))))......((((...))))........------ ( -18.10, z-score = -2.53, R) >droEre2.scaffold_4820 6943562 91 - 10470090 AAUAUUUCACUGCCCCAUGUGCAUGACUAAAGCCAUGCAUGUUUAUCCAUGUGAAAAAA---CUAUAUCAACAUCGUAUAAAAUACAAUCUCUC------ ....((((((((....(((((((((........))))))))).....)).))))))...---.............((((...))))........------ ( -15.50, z-score = -2.64, R) >droAna3.scaffold_13340 17306561 93 + 23697760 GAUUUUCAGCAGCUCCAAGUGCAUGCCUAGAGCCGUGCAUGUUUAUCUAUAGAAAAAAAUAACUAUAUCAACAUCGAAUUAAACACAGACUCU------- ....(((.(((((((...((....))...))))..)))(((((....(((((..........)))))..))))).)))...............------- ( -10.80, z-score = 0.71, R) >droWil1.scaffold_181089 9211306 96 + 12369635 CAAUUUUUACCGACGCUUGUACAUAUGCAUGUACAUGUACAACUGCAU-UGCAUGAUUGUUUUUCUAUCAAC-UAAACUAAACAAUCUUCUCUUAUUG-- ..........(((.((((((((((.((....)).))))))))..)).)-))...((((((((..........-......))))))))...........-- ( -14.99, z-score = -1.97, R) >consensus GAUAUUUCACUGCCCCAUGUGCAUGACUAAAGCCAUGCAUGUUUAUCUAUGUGAAAAAA___CUAUAUCAACAUCGUAUAAAAUACAAUCUCUC______ ....((((((......(((((((((.(....).)))))))))........))))))............................................ (-10.52 = -10.44 + -0.08)


| Location | 24,500,532 – 24,500,629 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 70.44 |
| Shannon entropy | 0.55935 |
| G+C content | 0.34526 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -10.48 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
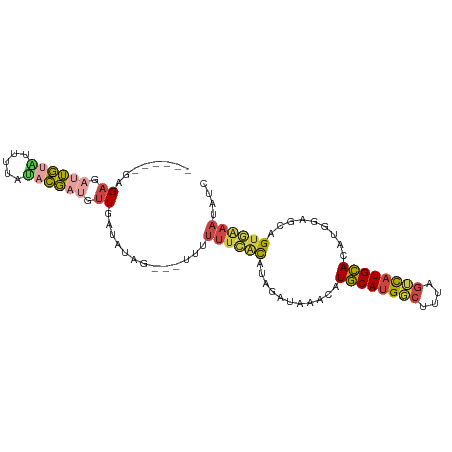
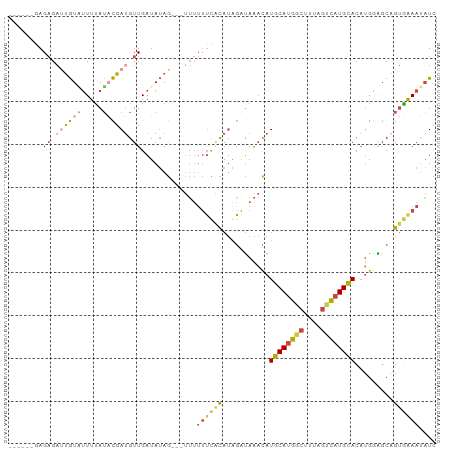
>dm3.chr3R 24500532 97 - 27905053 GGAAGAGAGAGAUUGUUUUUUAUACGAUGUUGAUAUAG---UUUGUUCACAUAGAUAAACAUGCAUGGCUUUAGUCAUGCACAUGGAGCAGUGAAAUAUC ...........((((((((((((...((((.((((...---..)))).))))..)))))..((((((((....))))))))....)))))))........ ( -21.50, z-score = -1.31, R) >droSim1.chr3R 24197959 97 - 27517382 GGAAGAGAGUGAUUGUAUUUUAUACGAUGUUGAUAUAG---UUUUUUCACAUAGAUAAACAUGCAUGGCUUUAGUCAUGCACAUGGAGCAGUGAAAUAUC ....(((((.(((((((((............)))))))---))))))).....((((....((((((((....))))))))(((......)))...)))) ( -19.70, z-score = -0.63, R) >droYak2.chr3R 6842571 91 + 28832112 ------GAGAGAUUGUAUUUUAUACGAUGUUGAUAUAG---UUUUUUCACAUGGCUAAACAUGCAUGGCUUUAGUCAUGCACAUGAGGCAGUGAAAUAUC ------..(((((((((((............)))))))---))))(((((.((.((.....((((((((....))))))))....)).)))))))..... ( -25.70, z-score = -2.82, R) >droEre2.scaffold_4820 6943562 91 + 10470090 ------GAGAGAUUGUAUUUUAUACGAUGUUGAUAUAG---UUUUUUCACAUGGAUAAACAUGCAUGGCUUUAGUCAUGCACAUGGGGCAGUGAAAUAUU ------..(((((((((((............)))))))---))))((((((((......)))(((((((....)))))))..........)))))..... ( -21.90, z-score = -2.04, R) >droAna3.scaffold_13340 17306561 93 - 23697760 -------AGAGUCUGUGUUUAAUUCGAUGUUGAUAUAGUUAUUUUUUUCUAUAGAUAAACAUGCACGGCUCUAGGCAUGCACUUGGAGCUGCUGAAAAUC -------((((.((((((........(((((..(((((..(....)..)))))....))))))))))))))).((((.((.......))))))....... ( -21.50, z-score = -0.84, R) >droWil1.scaffold_181089 9211306 96 - 12369635 --CAAUAAGAGAAGAUUGUUUAGUUUA-GUUGAUAGAAAAACAAUCAUGCA-AUGCAGUUGUACAUGUACAUGCAUAUGUACAAGCGUCGGUAAAAAUUG --...........((((((((..((((-.....)))).)))))))).(((.-((((..((((((((((......))))))))))))))..)))....... ( -21.80, z-score = -2.01, R) >consensus ______GAGAGAUUGUAUUUUAUACGAUGUUGAUAUAG___UUUUUUCACAUAGAUAAACAUGCAUGGCUUUAGUCAUGCACAUGGAGCAGUGAAAUAUC ........((.((((((.....)))))).)).............((((((...........((((((((....)))))))).........)))))).... (-10.48 = -11.07 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:59 2011