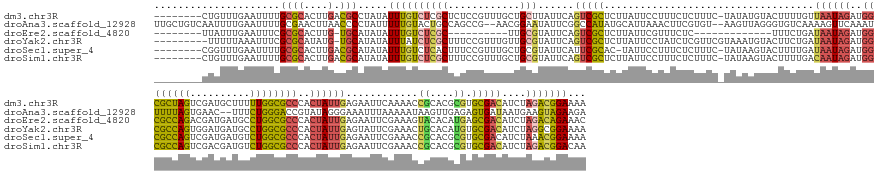
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,498,073 – 24,498,256 |
| Length | 183 |
| Max. P | 0.924146 |

| Location | 24,498,073 – 24,498,256 |
|---|---|
| Length | 183 |
| Sequences | 6 |
| Columns | 192 |
| Reading direction | forward |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.56485 |
| G+C content | 0.42232 |
| Mean single sequence MFE | -47.10 |
| Consensus MFE | -23.12 |
| Energy contribution | -24.80 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
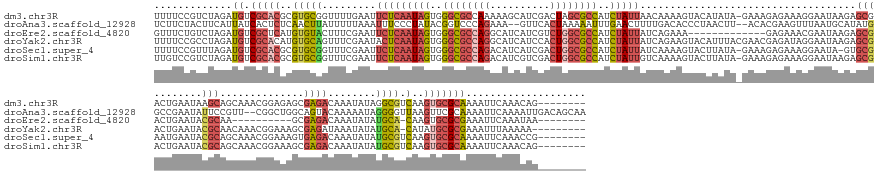
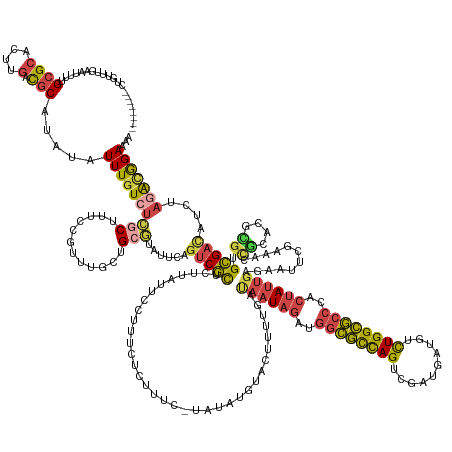
>dm3.chr3R 24498073 183 + 27905053 UUUUCCGUCUAGAUGUCGCACGCGUGCGGUUUUGAAUUCUCAAUAGUGGGCGCCAAAAAGCAUCGACUAGCGCCAUCUAUUAACAAAAGUACAUAUA-GAAAGAGAAAGGAAUAAGAGCGACUGAAUAAGCAGCAAACGGAGAGCGAGACAAAUAUAGGCGUCAAGUGCGCAAAAUUCAAACAG-------- ((((.(((((.(((((((((....))).((((((...(((((((((..(((((................)))))..)))))................-...................((..((.....))..)).....)))).)))))).......)))))).)).))).)))).........-------- ( -40.49, z-score = 0.41, R) >droAna3.scaffold_12928 102877 186 - 771024 UCUUCUACUUCAUUAUCACUCUCAACUUAUUUUUAAAUUUCCCUAUACGGUCCCAGAAA--GUUCACUAAAAAUUUGAACUUUUGACACCCUAACUU--ACACGAAGUUUAAUGCAUAUGGCCGAAUAUUCCGUU--CGGCUGGCAGUACAAAAAUAGGGGUUAAGUUCGCAAAAUUCAAAAUUGACAGCAA .......................((((((..........(((((((..(((....((((--(((((.........)))))))))...))).......--.......((....(((....((((((((.....)))--))))).)))..))....))))))).)))))).((.....(((....)))..)).. ( -35.30, z-score = -1.38, R) >droEre2.scaffold_4820 6941173 160 - 10470090 GUUUCUGUCUAGAUGUCGCUCAUGUGUACUUUCGAAUUCUCAAUAGUGGGCGCCAGGCAUCAUCGUCUGGCGCCAUCUAUUAUCAGAAA-------------GAGAAACGAAUAAGAGCGACUGAAUACGCAA----------GCGAGACAAAUAUAUGCA-CAAGUGCGCGAAAUUCAAAUAA-------- ((((((((((....(((((((.(((....((((...((((.(((((..((((((((((......))))))))))..)))))...)))).-------------..))))...))).)))))))......((...----------.))))))).......(((-....)))..)))))........-------- ( -54.10, z-score = -3.85, R) >droYak2.chr3R 6840131 182 - 28832112 UUUUCCGCCUAGAUGUCGCACAUGUGCAGUUUCGAAUACUCAAUAGUGGGCGCCAGGCAUCAUCCACUGGCGCCAUCUAUUAUCAGAAGUACAUUUACGAACGAGAUAGGAAUAAGAGCGACUGAAUACGCAACAAACGGAAAGCGAGAUAAAUAUAUGCA-CAUAUGCGCGAAAUUUAAAAA--------- .........(((((.((((.((((((((((((((....((((((((..((((((((..........))))))))..))))).......(((....)))....)))............(((........))).............)))))).......))))-))..)).)))).)))))....--------- ( -48.61, z-score = -2.03, R) >droSec1.super_4 3380207 182 + 6179234 UUUUCCGUUUAGAUGUCGCACGCGUGCGGUUUCGAAUUCUCAAUAGUGGGCGCCAGACAUCAUCGACUGGCGCCAUCUAUUAUCAAAAGUACUUAUA-GAAAGAGAAAGGAAUA-GUGCGAAUGAAUACGCAGCAAACGGAAAGUGAGACAAAUAUAUGCGUCAAGUGCGCAAAAUUCAAACCG-------- ((((((((((...(((..((..((..(.(((((...(((((.......((((((((.(......).)))))))).(((((..............)))-))..))))).))))).-)..))..)).....)))..)))))))))).............(((((.....)))))............-------- ( -52.84, z-score = -2.15, R) >droSim1.chr3R 24195554 183 + 27517382 UUGUCCGUCUAGAUGUCGCACGCGUGCGGUUUCGAAUUCUCAAUAGUGGGCGCCAGACAUCGUCGACUGGCGCCAUCUAUUGUCAAAAGUACUUAUA-GAAAGAGAAAGGAAUAAGAGCGACUGAAUACGCAGCAAACGGAAAGCGAGACAAAUAUAUGCGUCAAGUGCGCAAAAUUCAAACAG-------- ((((.((.((.(((((((((....))))((((((..((((((((((..((((((((.(......).))))))))..))))))......((.(((((.-.............))))).(((........))).))....))))..))))))........))))).)))).))))...........-------- ( -51.24, z-score = -0.80, R) >consensus UUUUCCGUCUAGAUGUCGCACGCGUGCGGUUUCGAAUUCUCAAUAGUGGGCGCCAGACAUCAUCGACUGGCGCCAUCUAUUAUCAAAAGUACAUAUA_GAAAGAGAAAGGAAUAAGAGCGACUGAAUACGCAGCAAACGGAAAGCGAGACAAAUAUAUGCGUCAAGUGCGCAAAAUUCAAACAG________ .............((.(((((...((((.........(((((((((..((((((((..........))))))))..)))))....................................(((........)))..............))))........))))....))))))).................... (-23.12 = -24.80 + 1.67)
| Location | 24,498,073 – 24,498,256 |
|---|---|
| Length | 183 |
| Sequences | 6 |
| Columns | 192 |
| Reading direction | reverse |
| Mean pairwise identity | 69.99 |
| Shannon entropy | 0.56485 |
| G+C content | 0.42232 |
| Mean single sequence MFE | -48.07 |
| Consensus MFE | -23.11 |
| Energy contribution | -24.79 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.919913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
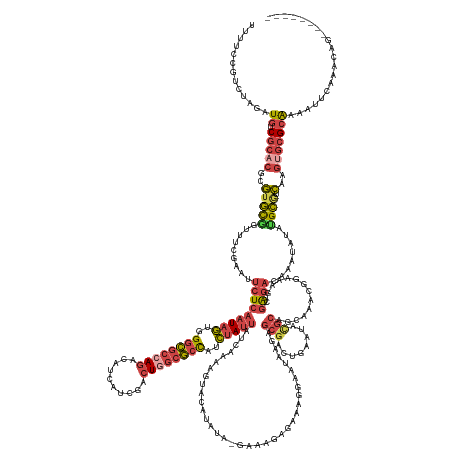
>dm3.chr3R 24498073 183 - 27905053 --------CUGUUUGAAUUUUGCGCACUUGACGCCUAUAUUUGUCUCGCUCUCCGUUUGCUGCUUAUUCAGUCGCUCUUAUUCCUUUCUCUUUC-UAUAUGUACUUUUGUUAAUAGAUGGCGCUAGUCGAUGCUUUUUGGCGCCCACUAUUGAGAAUUCAAAACCGCACGCGUGCGACAUCUAGACGGAAAA --------......((((...(((.(((.((((........))))..((.........)).........))))))....)))).....(((.((-((.((((....((.(((((((..((((((((..........))))))))..))))))).))........((((....)))))))).)))).)))... ( -41.50, z-score = -0.42, R) >droAna3.scaffold_12928 102877 186 + 771024 UUGCUGUCAAUUUUGAAUUUUGCGAACUUAACCCCUAUUUUUGUACUGCCAGCCG--AACGGAAUAUUCGGCCAUAUGCAUUAAACUUCGUGU--AAGUUAGGGUGUCAAAAGUUCAAAUUUUUAGUGAAC--UUUCUGGGACCGUAUAGGGAAAUUUAAAAAUAAGUUGAGAGUGAUAAUGAAGUAGAAGA (..((.((((((((((((((((((((..((.....))..)))))....((.((((--((.......))))))..(((((....(((((.....--)))))..(((.(((((((((((.........)))))--))).))).)))))))))))))))))))).....))))).))..)............... ( -43.10, z-score = -0.93, R) >droEre2.scaffold_4820 6941173 160 + 10470090 --------UUAUUUGAAUUUCGCGCACUUG-UGCAUAUAUUUGUCUCGC----------UUGCGUAUUCAGUCGCUCUUAUUCGUUUCUC-------------UUUCUGAUAAUAGAUGGCGCCAGACGAUGAUGCCUGGCGCCCACUAUUGAGAAUUCGAAAGUACACAUGAGCGACAUCUAGACAGAAAC --------.............((((....)-))).....(((((((((.----------...))......(((((((...((((.(((((-------------........(((((..((((((((.(......).))))))))..))))))))))..)))).(....)..)))))))....)))))))... ( -49.10, z-score = -2.75, R) >droYak2.chr3R 6840131 182 + 28832112 ---------UUUUUAAAUUUCGCGCAUAUG-UGCAUAUAUUUAUCUCGCUUUCCGUUUGUUGCGUAUUCAGUCGCUCUUAUUCCUAUCUCGUUCGUAAAUGUACUUCUGAUAAUAGAUGGCGCCAGUGGAUGAUGCCUGGCGCCCACUAUUGAGUAUUCGAAACUGCACAUGUGCGACAUCUAGGCGGAAAA ---------........(((((((((((((-(((((((((((((..((......(..((..(((........)))...))..)......))...))))))))).......((((((..((((((((.(.......)))))))))..))))))............)))))))))))(....)...)))))).. ( -51.80, z-score = -1.82, R) >droSec1.super_4 3380207 182 - 6179234 --------CGGUUUGAAUUUUGCGCACUUGACGCAUAUAUUUGUCUCACUUUCCGUUUGCUGCGUAUUCAUUCGCAC-UAUUCCUUUCUCUUUC-UAUAAGUACUUUUGAUAAUAGAUGGCGCCAGUCGAUGAUGUCUGGCGCCCACUAUUGAGAAUUCGAAACCGCACGCGUGCGACAUCUAAACGGAAAA --------.((..(((((..(((((....).))))...)))))..))..((((((((((.((((........)))).-..(((..(((((....-..((((....))))..(((((..((((((((.(......).))))))))..))))))))))...)))..((((....)))).....)))))))))). ( -50.80, z-score = -2.25, R) >droSim1.chr3R 24195554 183 - 27517382 --------CUGUUUGAAUUUUGCGCACUUGACGCAUAUAUUUGUCUCGCUUUCCGUUUGCUGCGUAUUCAGUCGCUCUUAUUCCUUUCUCUUUC-UAUAAGUACUUUUGACAAUAGAUGGCGCCAGUCGACGAUGUCUGGCGCCCACUAUUGAGAAUUCGAAACCGCACGCGUGCGACAUCUAGACGGACAA --------(((((((.((.((((((....((.(((......))).))...........((((((..(((....(..(((((.............-.)))))..)......((((((..((((((((.(......).))))))))..)))))).......)))..)))).)))))))).)).))))))).... ( -52.14, z-score = -1.77, R) >consensus ________CUGUUUGAAUUUUGCGCACUUGACGCAUAUAUUUGUCUCGCUUUCCGUUUGCUGCGUAUUCAGUCGCUCUUAUUCCUUUCUCUUUC_UAUAUGUACUUUUGAUAAUAGAUGGCGCCAGUCGAUGAUGUCUGGCGCCCACUAUUGAGAAUUCGAAACCGCACGCGUGCGACAUCUAGACGGAAAA ...........(((((((((((((.(((..(((((.........................)))))....))))))...................................((((((..((((((((..........))))))))..))))))))))))))))..((((....))))................ (-23.11 = -24.79 + 1.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:58 2011