
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,490,474 – 24,490,551 |
| Length | 77 |
| Max. P | 0.598847 |

| Location | 24,490,474 – 24,490,551 |
|---|---|
| Length | 77 |
| Sequences | 11 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.46467 |
| G+C content | 0.45185 |
| Mean single sequence MFE | -15.40 |
| Consensus MFE | -8.73 |
| Energy contribution | -9.27 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598847 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3R 24490474 77 - 27905053 -----CCGAAGCAACCCAAAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----...............((((.....((....-)).-.....(((((((((...........)))))))))......)))) ( -18.80, z-score = -2.08, R) >droSim1.chr3R 24187908 77 - 27517382 -----CCGAAGCAACCCAAAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----...............((((.....((....-)).-.....(((((((((...........)))))))))......)))) ( -18.80, z-score = -2.08, R) >droSec1.super_4 3372654 77 - 6179234 -----CCGAAGCAACCCAAAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----...............((((.....((....-)).-.....(((((((((...........)))))))))......)))) ( -18.80, z-score = -2.08, R) >droYak2.chr3R 6833817 77 + 28832112 -----CCGAAGCAACCCAAAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----...............((((.....((....-)).-.....(((((((((...........)))))))))......)))) ( -18.80, z-score = -2.08, R) >droEre2.scaffold_4820 6929647 77 + 10470090 -----CCGAAGCAACCCAAAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----...............((((.....((....-)).-.....(((((((((...........)))))))))......)))) ( -18.80, z-score = -2.08, R) >droAna3.scaffold_13340 17297946 76 - 23697760 -----CCAAGGCAACCC-AAAGCCGAAACGCACAG-GCA-AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----.((((.......-...(((..........)-)).-.....(((((((((...........))))))))).))))..... ( -19.70, z-score = -1.81, R) >dp4.chr2 27264575 78 + 30794189 -----CCGAAACGAAACCAAAACCGAAACGCACUG-GCAGAGCCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----........................((.(((-((...))).(((((((((...........))))))))).....)))). ( -16.00, z-score = -1.23, R) >droPer1.super_7 814993 78 - 4445127 -----CCGAAACGAAACCAAAACCGAAACGCACUG-GCAGAGCCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU -----........................((.(((-((...))).(((((((((...........))))))))).....)))). ( -16.00, z-score = -1.23, R) >droWil1.scaffold_181089 9199135 66 - 12369635 ----------------CAAAAAUUGUUGCACACAA-GUA-AAACCACAAAUUGCAUACAAAUGAAGCAAUUUACACUUGAUGCU ----------------...........(((..(((-((.-.......(((((((...........)))))))..))))).))). ( -8.40, z-score = -0.08, R) >droMoj3.scaffold_6540 29472235 64 - 34148556 --------------------AGGUGGAACGUACAGCUCAGCAACCGCCAAUUGCAUACAAAUGAACCAAUUUGCACUUGAGACG --------------------.(((((...((........))..))))).........(((((......)))))........... ( -9.40, z-score = 0.68, R) >droGri2.scaffold_14830 5464797 84 + 6267026 CCAAAAUAAGCCAUAAAUGUAAACGUAACGAAUCAAACCACAUCCACAAAUUGCAUUCAUAUGAACCAAUUUGCACUUGAGACU ........................................((....(((((((..(((....))).)))))))....))..... ( -5.90, z-score = 0.26, R) >consensus _____CCGAAGCAACCCAAAAGCCGAAACGCACAG_GCA_AACCCGCAAAUUGCAUACAAAUGAAGCAAUUUGCACUUGAGGCU .............................................(((((((((...........))))))))).......... ( -8.73 = -9.27 + 0.55)
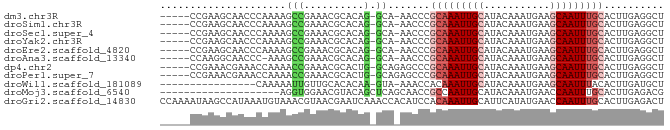

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:55 2011