
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,474,234 – 24,474,330 |
| Length | 96 |
| Max. P | 0.851847 |

| Location | 24,474,234 – 24,474,330 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.71 |
| Shannon entropy | 0.54790 |
| G+C content | 0.53605 |
| Mean single sequence MFE | -19.37 |
| Consensus MFE | -12.54 |
| Energy contribution | -12.56 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.851847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
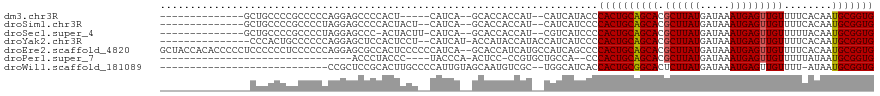
>dm3.chr3R 24474234 96 + 27905053 --------------GCUGCCCCGCCCCCAGGAGCCCCACU-----CAUCA--GCACCACCAU--CAUCAUACCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUCACAAUGCGGUG --------------((((...(.......)(((.....))-----)..))--))........--.........((((((((((.((((((.....)))))))))........))))))) ( -17.90, z-score = 0.05, R) >droSim1.chr3R 24171777 99 + 27517382 --------------GCUGCCCCGCCCCUAGGAGCCCCACUACU--CAUCA--GCACCACCAU--CAUCAUCCCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUCACAAUGCGGUG --------------((((...(.......)(((........))--)..))--))........--.........((((((((((.((((((.....)))))))))........))))))) ( -17.90, z-score = 0.01, R) >droSec1.super_4 3356546 99 + 6179234 --------------GCUGCCCCGCCCCUAGGAGCCC-ACUACUU-CAUCA--GCACCACCAU--CGUCAUCCCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUUACAAUGCGGUG --------------((((..((.......))((...-.))....-...))--))........--.........(((((((....((((((.....))))))(((....))).))))))) ( -17.70, z-score = 0.27, R) >droYak2.chr3R 6817288 101 - 28832112 ---------------CCCACUGCCCCCCAGGAGCUCCACUCCU--CAUCAU-ACCAUACCAUACCAUCAUCCCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUCACAAUGCGGUG ---------------..((((((.....(((((.....)))))--......-...........................((((.((((((.....))))))))))........)))))) ( -18.70, z-score = -1.42, R) >droEre2.scaffold_4820 6912191 117 - 10470090 GCUACCACACCCCCUCCCCCCUCCCCCCAGGAGCGCCACUCCCCCCAUCA--GCACCAUCAUGCCAUCAGCCCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUCACAAUGCGGUG (((..........................((((.....))))........--(((......)))....)))..((((((((((.((((((.....)))))))))........))))))) ( -20.80, z-score = -1.10, R) >droPer1.super_7 798993 79 + 4445127 --------------------------------ACCCUACCC----UACCCA-ACUCC-CCGUGCUGCCA--CCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUUAUAAUGCGGUG --------------------------------.........----((((((-.....-..(((((((..--......)))))))((((((.....))))))...........)).)))) ( -19.90, z-score = -3.81, R) >droWil1.scaffold_181089 9181061 88 + 12369635 ----------------------------CCGCUCCGCACUUGCCCCAUUGUAGCAAUGUCGC--UGGCAUCACCACUGCGGCACUCUUAUGAUAAAUGAGUUGUUUU-AUAAUGCGGUG ----------------------------.............(((.(((((((((((((((((--(((.....)))..))))))..(((((.....)))))))))..)-)))))).))). ( -22.70, z-score = -0.48, R) >consensus _______________CUGCCCCGCCCCCAGGAGCCCCACUCCU__CAUCA__GCACCACCAU__CAUCAUCCCCACUGCAGCACGCUUAUGAUAAAUGAGUUGUUUUCACAAUGCGGUG .........................................................................((((((((((.((((((.....)))))))))........))))))) (-12.54 = -12.56 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:53 2011