
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 814,638 – 814,786 |
| Length | 148 |
| Max. P | 0.856826 |

| Location | 814,638 – 814,746 |
|---|---|
| Length | 108 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.60441 |
| G+C content | 0.55080 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -15.98 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.856826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
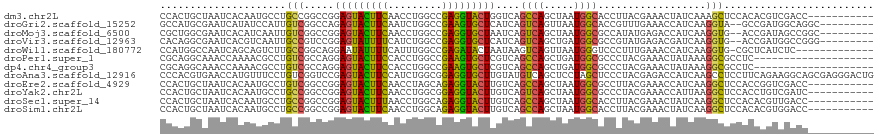
>dm3.chr2L 814638 108 - 23011544 CCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCCGAGGUACUGGUCAGCCAGCUAAUGGCACCUUACGAAACUAUCAAAGCUCCACACGUCGACC----------- .....(((.....((.(((((..(((((((.((......)))))))))))))).))....((((.....))))..................)))..............----------- ( -29.30, z-score = -0.87, R) >droGri2.scaffold_15252 7218102 108 - 17193109 GCCAUGCGAAUCAUAUCCAUUGUCGGCCAGAGUACUUCAAUCUGGCCGAAGUGCUCAUCAGUCAGUUAAUGGCACCGUUUGAAACCAUCAAGGUA--GCCGAUGGCAGGC--------- (((.(((..(((.....((((.(((((((((.........))))))))))))).................((((((...(((.....))).))).--)))))).))))))--------- ( -36.90, z-score = -2.69, R) >droMoj3.scaffold_6500 8486987 108 + 32352404 CGCUGGCGAAUCACAUCAAUUGUCGGCCGGAGUACUUCAACCUGGCCGAGGUGCUAAUCAGUCAGCUAAUGGCGCCAUAUGAGACCAUCAAGGUG--ACCGAUAGCCGGC--------- .((((((..(((...(((....((((((((.((......))))))))))(((((((...((....))..)))))))...))).(((.....))).--...))).))))))--------- ( -42.30, z-score = -2.79, R) >droVir3.scaffold_12963 19343863 108 - 20206255 CACAGGCGAAUCACGUCAAUUGCCGUCCGGAGUAUUUUCAUCUGGCCGAGGUGCUCAUCAGUCAGCUGAUGGCGCCGUAUGAGACGAUCAAGGUG--ACCGAUGGCCGGG--------- ....(((..(((.((((...(((((.(((((.........))))).)).(((((.((((((....))))))))))))))...)))).....(...--.).))).)))...--------- ( -41.00, z-score = -1.47, R) >droWil1.scaffold_180772 2678941 105 - 8906247 CCAUGGCCAAUCAGCAGUCUUGCCGGCAGGAAUAUUUUCAUUUGGCCGAGAUACUAAUAAGUCAGUUAAUGGGUCCCUUUGAAACCAUCAAGGUG-CGCUCAUCUC------------- ....((((((.......(((((....)))))..........))))))(((((..((((......))))..((((.(((((((.....)))))).)-.)))))))))------------- ( -26.83, z-score = -0.67, R) >droPer1.super_1 7098321 99 - 10282868 CGCAGGCAAACCAAAACGCCUGUCGCCAGGAGUACUUCCACCUGGCCGAAGUGCUCGUCAGCCAGCUGAUGGCGCCCUACGAAACUAUAAAGGCGCCUC-------------------- .((((((..........))))))......(((((((((.........)))))))))(((((....)))))((((((...............))))))..-------------------- ( -36.46, z-score = -2.42, R) >dp4.chr4_group3 5606161 99 - 11692001 CGCAGGCAAACCAAAACGCCUGUCGCCAGGAGUACUUCCACCUGGCCGAAGUGCUCGUCAGCCAGCUGAUGGCGCCCUACGAAACUAUAAAGGCGCCUC-------------------- .((((((..........))))))......(((((((((.........)))))))))(((((....)))))((((((...............))))))..-------------------- ( -36.46, z-score = -2.42, R) >droAna3.scaffold_12916 2704919 119 - 16180835 CCCACGUGAACCAUGUUUCCUGUCGGUCCGAGUACUUCCAUCUGGCGGAGGUGCUUGUAUGUCAGCUCCUAGCUCCCUACGAGACCAUCAAGCCUCCUUCAGAAGGCAGCGAGGGACUG ((((((((...))))).(((((((((...........)).(((((.((((((.((((((.(..(((.....)))..)))))))........)))))).))))).))))).))))).... ( -39.00, z-score = -0.69, R) >droEre2.scaffold_4929 870766 108 - 26641161 CCACUGCUAAUCACAAUGCCUGUCGGCCGGAGUACUUCAACCUAGCAGAGGUACUUGUCAGCCAGCUAAUGGCGCCUUACGAAACCAUCAAGGCUCCACCGGUCGACC----------- .....................(((((((((((((((((.........)))))))).....((((.....))))(((((..((.....)))))))....))))))))).----------- ( -36.60, z-score = -2.67, R) >droYak2.chr2L 798207 108 - 22324452 CCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCGGAGGUACUUGUCAGUCAGCUAAUGGCGCCCUACGAAACCAUUAAGGCUCCACCUGUCGAUC----------- .........((((((((((((.(((.((((.((......)))))))))))))).))))..(.(((....(((.(((...............))).))).))).)))).----------- ( -28.56, z-score = 0.24, R) >droSec1.super_14 785007 108 - 2068291 CCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUUAACCUGGCAGAGGUACUUGUCAGCCAGCUAAUGGCACCUUACGAAACUAUCAAGGCUCCACACGUUGACC----------- .....(((....(((((((((....(((((.((......)))))))..))))).)))).)))((((...(((..((((..((.....))))))..)))...))))...----------- ( -25.80, z-score = 0.37, R) >droSim1.chr2L 816781 108 - 22036055 CCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCAGAGGUACUUGUCAGCCAGCUAAUGGCACCUUACGAAACUAUCAAGGCUCCACACGUGGACC----------- ((((............((((.((.(((.((((((((((.........)))))))))..).))).))....))))((((..((.....))))))........))))...----------- ( -29.10, z-score = -0.17, R) >consensus CCAAUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCCGAGGUACUUGUCAGCCAGCUAAUGGCGCCCUACGAAACCAUCAAGGCUCCACCCGUCGACC___________ .....................(((.....(((((((((.........)))))))))....((((.....))))..................)))......................... (-15.98 = -14.98 + -0.99)

| Location | 814,666 – 814,786 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.07 |
| Shannon entropy | 0.50044 |
| G+C content | 0.55903 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -22.25 |
| Energy contribution | -20.93 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
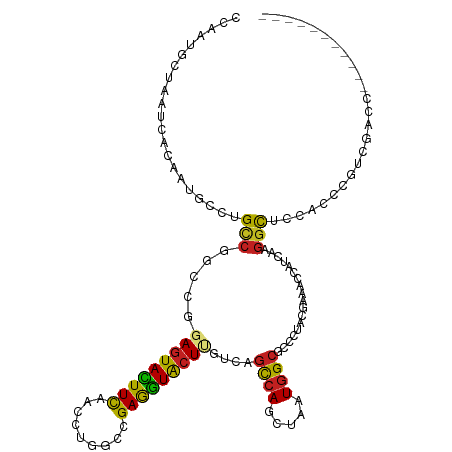
>dm3.chr2L 814666 120 - 23011544 CAGCGAUGCUGGCGACUGCUCCGAGCAUGCAACUACAACGCCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCCGAGGUACUGGUCAGCCAGCUAAUGGCACCUUAC ..((.(((((((((((.......((((.((.........))...)))).....((.(((((..(((((((.((......)))))))))))))).))))).))))))..)).))....... ( -42.00, z-score = -1.42, R) >droGri2.scaffold_15252 7218130 120 - 17193109 CAACGCUGCUGGCGUCUGUUGCGCGCCUGUAAUUACAAUGGCCAUGCGAAUCAUAUCCAUUGUCGGCCAGAGUACUUCAAUCUGGCCGAAGUGCUCAUCAGUCAGUUAAUGGCACCGUUU .((((.(((((((..(((.((.((((.........((((((..(((.....)))..))))))(((((((((.........))))))))).)))).)).)))...)))...)))).)))). ( -42.60, z-score = -2.60, R) >droMoj3.scaffold_6500 8487015 120 + 32352404 CAGCGCUGCUGGCGAUUACUGCGCGCUUGUAACUACAAUCCGCUGGCGAAUCACAUCAAUUGUCGGCCGGAGUACUUCAACCUGGCCGAGGUGCUAAUCAGUCAGCUAAUGGCGCCAUAU ..((((((((((((((((..(((((((.((...........)).)))...............((((((((.((......)))))))))).))))))))).))))))....)))))..... ( -43.40, z-score = -1.65, R) >droVir3.scaffold_12963 19343891 120 - 20206255 CAACGCUGUUGGCGCCUGUUGCGCGCCUGUAAUUACAACACACAGGCGAAUCACGUCAAUUGCCGUCCGGAGUAUUUUCAUCUGGCCGAGGUGCUCAUCAGUCAGCUGAUGGCGCCGUAU ....((.(((((((..((.....(((((((...........)))))))...))))))))).))((.(((((.........))))).)).(((((.((((((....))))))))))).... ( -46.40, z-score = -2.28, R) >droWil1.scaffold_180772 2678966 120 - 8906247 CAACGGUGCUGGCGUUUGUUAAGGGCCUGCAAUUUCAAUCCCAUGGCCAAUCAGCAGUCUUGCCGGCAGGAAUAUUUUCAUUUGGCCGAGAUACUAAUAAGUCAGUUAAUGGGUCCCUUU ....(((((.(((.((.....)).))).)))........(((((((((((.......(((((....)))))..........))))))..(((........))).....)))))..))... ( -31.23, z-score = 0.36, R) >droPer1.super_1 7098340 120 - 10282868 CGACGUUGCUGGCGGCUGCUCCGCGCCUGUAACUACAAUUCGCAGGCAAACCAAAACGCCUGUCGCCAGGAGUACUUCCACCUGGCCGAAGUGCUCGUCAGCCAGCUGAUGGCGCCCUAC ....(((((.((((.(......))))).))))).....(((((((((..........)))))).((((((.(......).)))))).)))((((.((((((....))))))))))..... ( -46.50, z-score = -1.85, R) >dp4.chr4_group3 5606180 120 - 11692001 CGACGUUGCUGGCGGCUGCUCCGCGCCUGUAACUACAAUCCGCAGGCAAACCAAAACGCCUGUCGCCAGGAGUACUUCCACCUGGCCGAAGUGCUCGUCAGCCAGCUGAUGGCGCCCUAC .(((....((((((((.((...))((((((...........))))))..............))))))))(((((((((.........)))))))))))).((((.....))))....... ( -45.90, z-score = -1.67, R) >droAna3.scaffold_12916 2704958 120 - 16180835 CAGCGUUGCUGGCGACUCCUGCGCGCCUGCAACUACAAUUCCCACGUGAACCAUGUUUCCUGUCGGUCCGAGUACUUCCAUCUGGCGGAGGUGCUUGUAUGUCAGCUCCUAGCUCCCUAC .((((((((.((((.(......))))).)))))..........(((((...)))))...(((.((...(((((((((((.......)))))))))))..)).)))......)))...... ( -37.60, z-score = -1.08, R) >droEre2.scaffold_4929 870794 120 - 26641161 CAGAGAUGCUGGCGACUGCUCCGUGCAUGCAACUACAACGCCACUGCUAAUCACAAUGCCUGUCGGCCGGAGUACUUCAACCUAGCAGAGGUACUUGUCAGCCAGCUAAUGGCGCCUUAC .......((((((((((((((((((...(((.............)))....)))...(((....))).)))))).....((((.....))))....))).)))))).............. ( -34.02, z-score = 0.64, R) >droYak2.chr2L 798235 120 - 22324452 CAGAGAUGCUGGCGACUGCUCCGUGCAUGCAACUACAACGCCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCGGAGGUACUUGUCAGUCAGCUAAUGGCGCCCUAC ..........((((..(((.....)))..).........((((..(((.((.(((((((((.(((.((((.((......)))))))))))))).))))..)).)))...))))))).... ( -36.30, z-score = 0.57, R) >droSec1.super_14 785035 120 - 2068291 CAGCGAUGCUGGCGACUGCUCCGAGCAUGCAACUACAACGCCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUUAACCUGGCAGAGGUACUUGUCAGCCAGCUAAUGGCACCUUAC ..((.((((((((((((((((((((((.((.........))...)))).........(((....)))))))))).....((((.....))))....))).))))))..)).))....... ( -38.60, z-score = -0.86, R) >droSim1.chr2L 816809 120 - 22036055 CAGCGAUGCUGGCGACUGCUUCGAGCAUGCAACUACAACGCCACUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCAGAGGUACUUGUCAGCCAGCUAAUGGCACCUUAC ..((.((((((((((((((((((.(((.(((........((....)).........))).)))).(((((.((......))))))).))))))...))).))))))..)).))....... ( -38.23, z-score = -0.70, R) >consensus CAGCGAUGCUGGCGACUGCUCCGCGCCUGCAACUACAACGCCAAUGCUAAUCACAAUGCCUGCCGGCCGGAGUACUUCAACCUGGCCGAGGUACUUGUCAGCCAGCUAAUGGCGCCCUAC .......((((((..........((((.................)))).....................(((((((((.........)))))))))....)))))).............. (-22.25 = -20.93 + -1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:07:02 2011