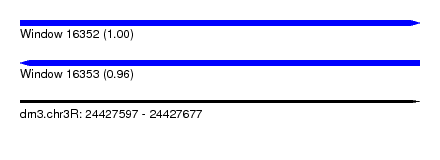
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,427,597 – 24,427,677 |
| Length | 80 |
| Max. P | 0.999610 |

| Location | 24,427,597 – 24,427,677 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 67.09 |
| Shannon entropy | 0.71158 |
| G+C content | 0.46453 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -12.38 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
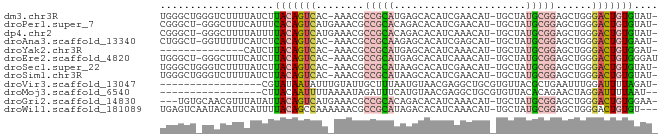
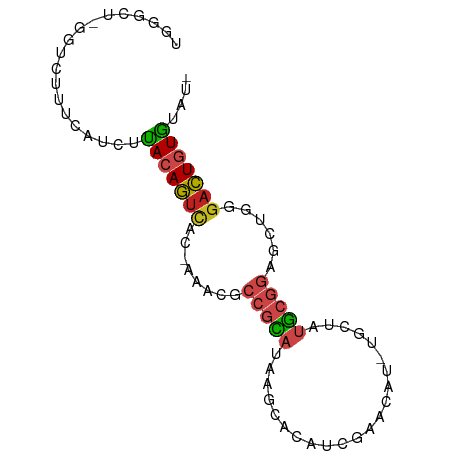
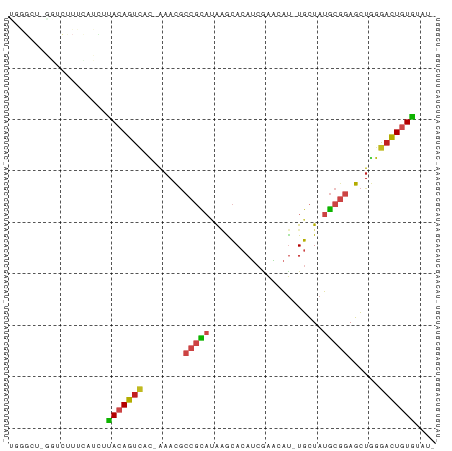
>dm3.chr3R 24427597 80 + 27905053 -AUACACAGUCCCAGCUCCGCAUAGCA-AUGUUCGAUGUGCUCAUGCGGCGUUU-GUGACUGUAAGAUAAAAGACCCAGCCCA -....(((((((.(((.((((((((((-..........)))).)))))).))).-).)))))).................... ( -24.20, z-score = -2.59, R) >droPer1.super_7 736040 80 + 4445127 -AUACACAGUCCCAGCUCCGCAUAGCA-AUGUUCGAUGUGUCUGUGCGGCGUUUCAUGACUGUGAAAUGAAAGCCC-AGCCCG -...(((((((..(((.((((((((((-..........)).)))))))).)))....)))))))............-...... ( -25.30, z-score = -2.54, R) >dp4.chr2 27181174 80 - 30794189 -AUACACAGUCCCAGCUCCGCAUAGCA-AUGUUUGAUGUGUCUGUGCGGCGUUUCAUGACUGUAAAAUAAAAGCCC-AGCCCG -....((((((..(((.((((((((((-..........)).)))))))).)))....)))))).............-...... ( -22.10, z-score = -2.37, R) >droAna3.scaffold_13340 17232747 79 + 23697760 -AUUCACAGUCCCAGCUCCGCAUAGCA-AUGCUCGAUGUGCUCUUGCGGCGUUU-GUGACUGUGAGAUGAAAAACC-AGCCAG -.((((((((((.(((.(((((.((((-..........))))..))))).))).-).)))))))))..........-...... ( -28.00, z-score = -2.85, R) >droYak2.chr3R 6765887 66 - 28832112 -AUCCACAGUCCCAGCUCCGCAUAGCA-AUGUUUGAUGUGCUCAUGCGGCGUUU-GUGACUGUAAGAUG-------------- -(((.(((((((.(((.((((((((((-..........)))).)))))).))).-).))))))..))).-------------- ( -24.30, z-score = -3.29, R) >droEre2.scaffold_4820 6864107 80 - 10470090 AUCCCACAGUCCCAGCUCCGCAUAGCA-AUGUUUGAUGUGCUCAUGCGGCGUUU-GUGACUGUAAGAUGAAAGCCC-AGCCCA (((..(((((((.(((.((((((((((-..........)))).)))))).))).-).))))))..)))........-...... ( -25.50, z-score = -2.77, R) >droSec1.super_22 1029995 80 + 1066717 -AUACACAGUCCCAGCUCCGCAUAGCA-AUGUUCGAUGUGCUUAUGCGGCGUUU-GUGACUGUAAGAUAAAAGACCCAGCCCA -....(((((((.(((.((((((((((-..........))).))))))).))).-).)))))).................... ( -24.20, z-score = -2.76, R) >droSim1.chr3R 24118719 80 + 27517382 -AUACACAGUCCCAGCUCCGCAUAGCA-AUGUUCGAUGUGCUUAUGCGGCGUUU-GUGACUGUAAGAUAAAAGACCCAGCCCA -....(((((((.(((.((((((((((-..........))).))))))).))).-).)))))).................... ( -24.20, z-score = -2.76, R) >droVir3.scaffold_13047 6807236 65 - 19223366 -AUCUAAAAUCCAAAUUCAGCGUAACACGCAGCCUCGUUACAUUAAAGCAAUACAAAUAUUAUACG----------------- -..................(((.....)))....................................----------------- ( -4.00, z-score = -0.47, R) >droMoj3.scaffold_6540 29401129 64 + 34148556 --AUUAAAAUCCUAGUUCUGUGUAACACGCAGCCUCGUUACAUGAAAUCUAUUUUAAAAUUGUAAG----------------- --.(((((((.....(((.(((((((..........))))))))))....))))))).........----------------- ( -9.40, z-score = -1.93, R) >droGri2.scaffold_14830 715003 78 + 6267026 -UUCCACAGUCCCAGCUCCGCAUAGCA-AUGUUUGAUGUGUCUGUGCGGCGUUUCAUGACUGUAAUAUAAACGUUGCACA--- -....((((((..(((.((((((((((-..........)).)))))))).)))....)))))).................--- ( -22.10, z-score = -1.65, R) >droWil1.scaffold_181089 9124087 79 + 12369635 ---ACACAGUCCCAGCUCCGCAUAGCA-AUGUUUGAUGUGUCUAUGCGGCGUUUUUUGGCUGUAAAAUGAAUGUAUUGACUCA ---..((((((..(((.((((((((((-..........)).)))))))).)))....)))))).................... ( -21.50, z-score = -1.29, R) >consensus _AUACACAGUCCCAGCUCCGCAUAGCA_AUGUUCGAUGUGCUUAUGCGGCGUUU_GUGACUGUAAGAUAAAAGACC_AGCCCA .....((((((..(((.(((((..(((.((.....)).)))...))))).)))....)))))).................... (-12.38 = -13.20 + 0.82)

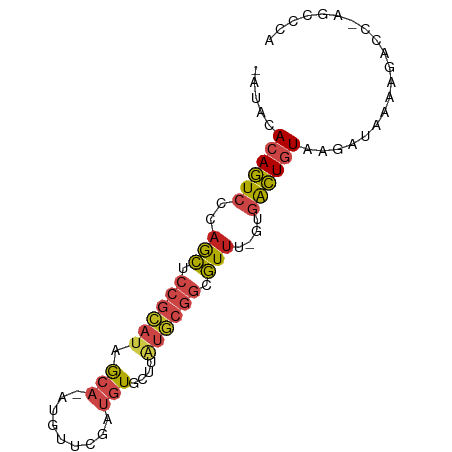
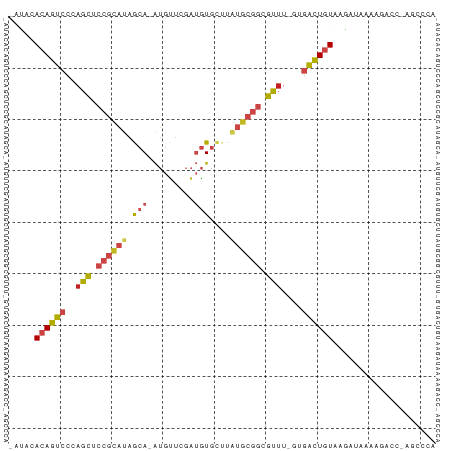
| Location | 24,427,597 – 24,427,677 |
|---|---|
| Length | 80 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 67.09 |
| Shannon entropy | 0.71158 |
| G+C content | 0.46453 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -10.75 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24427597 80 - 27905053 UGGGCUGGGUCUUUUAUCUUACAGUCAC-AAACGCCGCAUGAGCACAUCGAACAU-UGCUAUGCGGAGCUGGGACUGUGUAU- .((((...)))).......(((((((.(-(....((((((.((((..........-))))))))))...)).)))))))...- ( -23.00, z-score = -0.80, R) >droPer1.super_7 736040 80 - 4445127 CGGGCU-GGGCUUUCAUUUCACAGUCAUGAAACGCCGCACAGACACAUCGAACAU-UGCUAUGCGGAGCUGGGACUGUGUAU- .((((.-..))))......(((((((........(((((.((.((..........-)))).)))))......)))))))...- ( -19.34, z-score = 0.39, R) >dp4.chr2 27181174 80 + 30794189 CGGGCU-GGGCUUUUAUUUUACAGUCAUGAAACGCCGCACAGACACAUCAAACAU-UGCUAUGCGGAGCUGGGACUGUGUAU- .((((.-..))))......(((((((........(((((.((.((..........-)))).)))))......)))))))...- ( -17.84, z-score = 0.25, R) >droAna3.scaffold_13340 17232747 79 - 23697760 CUGGCU-GGUUUUUCAUCUCACAGUCAC-AAACGCCGCAAGAGCACAUCGAGCAU-UGCUAUGCGGAGCUGGGACUGUGAAU- ......-...........((((((((.(-(....(((((..((((..........-)))).)))))...)).))))))))..- ( -24.70, z-score = -1.05, R) >droYak2.chr3R 6765887 66 + 28832112 --------------CAUCUUACAGUCAC-AAACGCCGCAUGAGCACAUCAAACAU-UGCUAUGCGGAGCUGGGACUGUGGAU- --------------.((((.((((((.(-(....((((((.((((..........-))))))))))...)).))))))))))- ( -24.10, z-score = -3.53, R) >droEre2.scaffold_4820 6864107 80 + 10470090 UGGGCU-GGGCUUUCAUCUUACAGUCAC-AAACGCCGCAUGAGCACAUCAAACAU-UGCUAUGCGGAGCUGGGACUGUGGGAU .((((.-..))))..(((((((((((.(-(....((((((.((((..........-))))))))))...)).))))))))))) ( -28.50, z-score = -2.61, R) >droSec1.super_22 1029995 80 - 1066717 UGGGCUGGGUCUUUUAUCUUACAGUCAC-AAACGCCGCAUAAGCACAUCGAACAU-UGCUAUGCGGAGCUGGGACUGUGUAU- .((((...)))).......(((((((.(-(....(((((((.(((..........-))))))))))...)).)))))))...- ( -23.00, z-score = -1.14, R) >droSim1.chr3R 24118719 80 - 27517382 UGGGCUGGGUCUUUUAUCUUACAGUCAC-AAACGCCGCAUAAGCACAUCGAACAU-UGCUAUGCGGAGCUGGGACUGUGUAU- .((((...)))).......(((((((.(-(....(((((((.(((..........-))))))))))...)).)))))))...- ( -23.00, z-score = -1.14, R) >droVir3.scaffold_13047 6807236 65 + 19223366 -----------------CGUAUAAUAUUUGUAUUGCUUUAAUGUAACGAGGCUGCGUGUUACGCUGAAUUUGGAUUUUAGAU- -----------------.(..((((((..(((..(((((........))))))))))))))..)..................- ( -7.80, z-score = 1.15, R) >droMoj3.scaffold_6540 29401129 64 - 34148556 -----------------CUUACAAUUUUAAAAUAGAUUUCAUGUAACGAGGCUGCGUGUUACACAGAACUAGGAUUUUAAU-- -----------------.........(((((((..(.(((.(((((((........)))))))..))).)...))))))).-- ( -8.30, z-score = -0.39, R) >droGri2.scaffold_14830 715003 78 - 6267026 ---UGUGCAACGUUUAUAUUACAGUCAUGAAACGCCGCACAGACACAUCAAACAU-UGCUAUGCGGAGCUGGGACUGUGGAA- ---...............((((((((........(((((.((.((..........-)))).)))))......))))))))..- ( -18.44, z-score = -0.23, R) >droWil1.scaffold_181089 9124087 79 - 12369635 UGAGUCAAUACAUUCAUUUUACAGCCAAAAAACGCCGCAUAGACACAUCAAACAU-UGCUAUGCGGAGCUGGGACUGUGU--- (((((......)))))...(((((((........((((((((.((..........-)))))))))).....)).))))).--- ( -20.92, z-score = -2.20, R) >consensus UGGGCU_GGUCUUUCAUCUUACAGUCAC_AAACGCCGCAUAAGCACAUCGAACAU_UGCUAUGCGGAGCUGGGACUGUGUAU_ ...................(((((((........(((((......................)))))......))))))).... (-10.75 = -10.92 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:46 2011