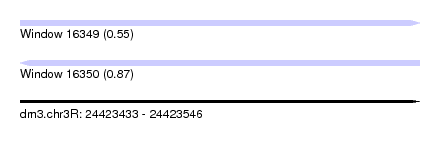
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,423,433 – 24,423,546 |
| Length | 113 |
| Max. P | 0.872222 |

| Location | 24,423,433 – 24,423,546 |
|---|---|
| Length | 113 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 95.94 |
| Shannon entropy | 0.08157 |
| G+C content | 0.37868 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.545825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
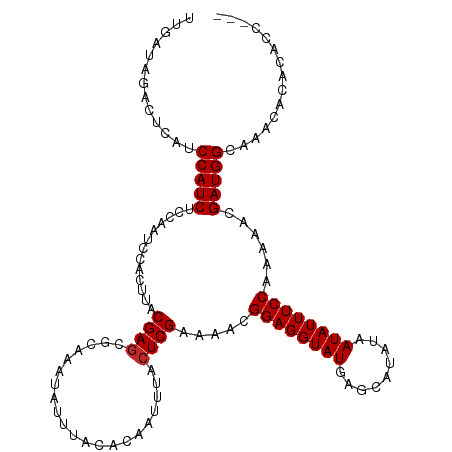
>dm3.chr3R 24423433 113 + 27905053 ---GUGGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAG ---.((((((((((.(((.......))))))))).....((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))..)))). ( -26.40, z-score = -0.81, R) >droPer1.super_0 4872663 114 + 11822988 --GUGUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGUUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAA --(((((.((((((.(((.......))))))))))))))(((((....(((((((((((..............(((((((....)))))))..))))))))))))))))....... ( -25.29, z-score = -0.94, R) >droAna3.scaffold_13340 17229183 112 + 23697760 ----GUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAA ----((((((((((.(((.......)))))))).)))))((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))....... ( -26.50, z-score = -1.24, R) >droEre2.scaffold_4820 6860260 113 - 10470090 ---GUGGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAA ---.((((((((((.(((.......))))))))).....((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))..)))). ( -26.40, z-score = -0.88, R) >droYak2.chr3R 6762084 115 - 28832112 -GUGGUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAA -..((((((.((...(((.......)))...........)).))))))(((((((.(((((.((.(((....))).))..))))).))))))).....((((((....)))))).. ( -27.34, z-score = -1.07, R) >droSec1.super_22 1026058 113 + 1066717 ---GUGGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAG ---.((((((((((.(((.......))))))))).....((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))..)))). ( -26.40, z-score = -0.81, R) >droSim1.chr3R 24114820 113 + 27517382 ---GUGGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAG ---.((((((((((.(((.......))))))))).....((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))..)))). ( -26.40, z-score = -0.81, R) >droMoj3.scaffold_6540 17188107 116 + 34148556 GUGUGUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUUUAUCAA ..(((((.((((((.(((.......))))))))))))))((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))....... ( -27.20, z-score = -1.28, R) >droGri2.scaffold_14906 1468830 116 + 14172833 GUGUGUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUUUAUCAA ..(((((.((((((.(((.......))))))))))))))((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....))))))....... ( -27.20, z-score = -1.28, R) >droVir3.scaffold_12822 3905757 116 + 4096053 GUGUCUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUUUAUCAA ........((((((.(((.......))))))))).(((.((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....)))))).)))... ( -25.80, z-score = -0.85, R) >consensus ___GGUGUGUGUUUGCCAUCGUUUUUGGAAAUAUUAUAUGCUCAUACCUCCGUUUUCGAGUAAAUUGUGUAAAUAUUUGCGCUCGUAAGUGGAUUGGAGAUGGAUGAGUCUAUCAA ........((((((.(((.......))))))))).(((.((((((..(((((..((((..((....((((((....))))))...))..)))).)))))....)))))).)))... (-25.44 = -25.35 + -0.09)



| Location | 24,423,433 – 24,423,546 |
|---|---|
| Length | 113 |
| Sequences | 10 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 95.94 |
| Shannon entropy | 0.08157 |
| G+C content | 0.37868 |
| Mean single sequence MFE | -15.92 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.88 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.872222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24423433 113 - 27905053 CUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACCAC--- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............--- ( -16.20, z-score = -1.19, R) >droPer1.super_0 4872663 114 - 11822988 UUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAACGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACACAC-- .............(((((.............(((.......................))).....((((((((.........))))))))......))))).............-- ( -13.40, z-score = -0.68, R) >droAna3.scaffold_13340 17229183 112 - 23697760 UUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACAC---- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))...........---- ( -16.20, z-score = -1.37, R) >droEre2.scaffold_4820 6860260 113 + 10470090 UUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACCAC--- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............--- ( -16.20, z-score = -1.24, R) >droYak2.chr3R 6762084 115 + 28832112 UUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACACCAC- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))..............- ( -16.20, z-score = -1.36, R) >droSec1.super_22 1026058 113 - 1066717 CUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACCAC--- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............--- ( -16.20, z-score = -1.19, R) >droSim1.chr3R 24114820 113 - 27517382 CUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACCAC--- .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............--- ( -16.20, z-score = -1.19, R) >droMoj3.scaffold_6540 17188107 116 - 34148556 UUGAUAAACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACACACAC .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............... ( -16.20, z-score = -1.83, R) >droGri2.scaffold_14906 1468830 116 - 14172833 UUGAUAAACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACACACAC .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............... ( -16.20, z-score = -1.83, R) >droVir3.scaffold_12822 3905757 116 - 4096053 UUGAUAAACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACAGACAC .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............... ( -16.20, z-score = -1.45, R) >consensus UUGAUAGACUCAUCCAUCUCCAAUCCACUUACGAGCGCAAAUAUUUACACAAUUUACUCGAAAACGGAGGUAUGAGCAUAUAAUAUUUCCAAAAACGAUGGCAAACACACACC___ .............(((((.............((((.....................)))).....((((((((.........))))))))......)))))............... (-15.78 = -15.88 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:44 2011