| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,410,315 – 24,410,412 |
| Length | 97 |
| Max. P | 0.950972 |

| Location | 24,410,315 – 24,410,412 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.55 |
| Shannon entropy | 0.63256 |
| G+C content | 0.51488 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -14.36 |
| Energy contribution | -13.94 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.950972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
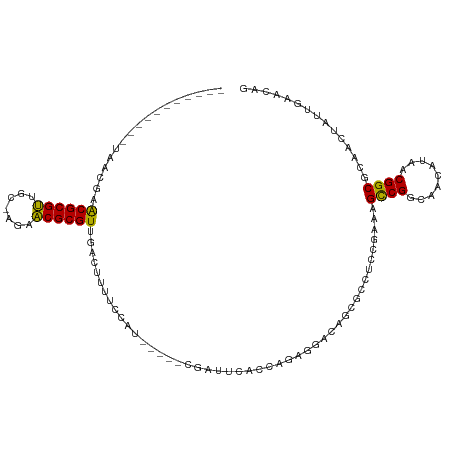
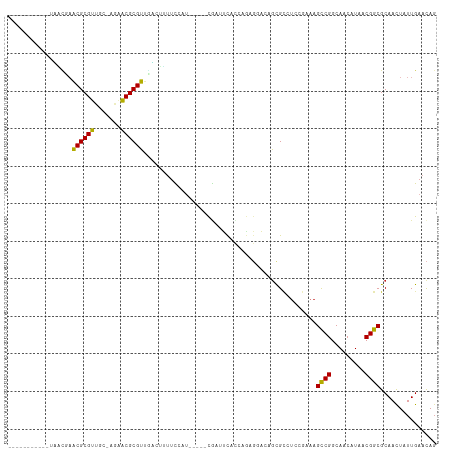
>dm3.chr3R 24410315 97 - 27905053 -----------UAACUAACGCGUUGC-AGAACGCGUUGCCUUUUCCAU-----CGAUCCACCAGAGGGCAGCGCCUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------..........(((((-.....((((((((((((....-----.........))))))))))))........((((.........))))))))).......... ( -31.22, z-score = -2.46, R) >droSim1.chr3R 24101849 97 - 27517382 -----------UGACUAACGCGUUGC-AGAACGCGUUGCCUUUUCCAU-----CGAUCCACCAGAGGGCAGCGCCUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------..........(((((-.....((((((((((((....-----.........))))))))))))........((((.........))))))))).......... ( -31.22, z-score = -2.05, R) >droSec1.super_22 1013063 97 - 1066717 -----------UGACUAACGCGUUGC-AGAACGCGUUGCCUUUUCCAU-----CGAUCCACCAGAGGGCAGCGCCUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------..........(((((-.....((((((((((((....-----.........))))))))))))........((((.........))))))))).......... ( -31.22, z-score = -2.05, R) >droYak2.chr3R 6747650 97 + 28832112 -----------UGACUAACGCGUUGC-AGAACGCGUUGCCUUUUCCAU-----CGAUCCACCAGAGGGCAGCGCCUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------..........(((((-.....((((((((((((....-----.........))))))))))))........((((.........))))))))).......... ( -31.22, z-score = -2.05, R) >droEre2.scaffold_4820 6847177 97 + 10470090 -----------UGACUAACGCGUUGC-AGAACGCGUUGCCUUUUCCAU-----CGAUCCACCAGAGGGCAGCGCCUCCGACAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------..........(((((-.....((((((((((((....-----.........))))))))))))........((((.........))))))))).......... ( -31.22, z-score = -2.11, R) >droAna3.scaffold_13340 17215836 86 - 23697760 ------------UGAAAACGCGUACC--GCACGCGU-UACCCUUUCCA-----UACAUCC--------ACACGAGUCCAAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG ------------....(((((((...--..))))))-)..........-----.......--------...(((........((((.........)))).......)))..... ( -16.16, z-score = -0.69, R) >dp4.chr2 15238151 98 + 30794189 -----------UUUUGUGCGCGUCCCUUGAACGCGUCAAACUUUCCCU-----CGCUUCUCCUGGGGACAGUGCGUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------(((((..(((((((((.(((.(((.............-----))))))....))))))...)))..)))))((((.........))))............... ( -29.12, z-score = -0.85, R) >droPer1.super_0 4857779 98 - 11822988 -----------UUUUGUGCGCGUCCCUUGCACGCGUCAAACUUUCCCU-----CGCUUCUCCUGGGGACAGUGCGUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG -----------....(((((.......)))))(((((...(((((...-----(((.....(((....))).)))...)))))..(....).....)))))............. ( -28.00, z-score = -0.32, R) >droWil1.scaffold_181089 9100565 91 - 12369635 -----------UCGUAAACGCGUUCCCGGAACGCGUGUGACUCCUCUU-----UACAUC-------GAACGCGUUCGAAAAAGCCGGCAACAUAACGGCGCAACUAUUGAAUAG -----------..((..((((((((...))))))))...)).......-----....((-------(((....)))))....((((.........))))............... ( -24.00, z-score = -0.71, R) >droVir3.scaffold_12822 3889776 108 - 4096053 UGUAUA-----AAUCGAACGCGUCGCUAGAACGCGUUUUAAAUUUAUUGCAUUUGCAUUUUAAACGCAGCGCGU-UAAAAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG ((((((-----(((.(((((((((....).))))))))...))))).)))).((((.((((.(((((...))))-).)))).((((.........))))))))........... ( -28.90, z-score = -1.12, R) >droMoj3.scaffold_6540 17172012 106 - 34148556 UUUGUA-----AAUCGAGCGCGCGGAUUAGGCGCGUCGACAAUUUCUUCCG--UAAAUUUUUAACGCAGCGCGU-UCAAAAAGUCGGCAACAUAACGGCGCAAUUAUUGAACAG .((((.-----....((((((((.(......)((((..(.((((((....)--.))))).)..)))).))))))-)).....((((.........))))))))........... ( -27.00, z-score = -0.08, R) >droGri2.scaffold_14906 1453640 111 - 14172833 UGUAUAAAAAUAAUUGAACGCGUCGAGAGAACGCGUUGAUUAAUUCUAGCA--UCAAUUUCAAACGCACCGCGU-UAAUAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAC (((.(((...(((((.((((((((....).))))))))))))......((.--.........(((((...))))-)......((((.........)))))).....))).))). ( -27.00, z-score = -1.86, R) >consensus ___________UAACGAACGCGUUGC_AGAACGCGUUGACUUUUCCAU_____CGAUUCACCAGAGGACAGCGCCUCCGAAAGCCGGCAACAUAACGGCGCAACUAUUGAACAG .................((((((.......))))))..............................................((((.........))))............... (-14.36 = -13.94 + -0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:41 2011