
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,394,201 – 24,394,309 |
| Length | 108 |
| Max. P | 0.666557 |

| Location | 24,394,201 – 24,394,309 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.67 |
| Shannon entropy | 0.60361 |
| G+C content | 0.50323 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -16.95 |
| Energy contribution | -18.87 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.61 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
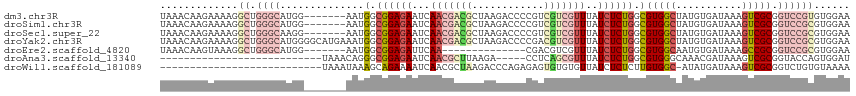
>dm3.chr3R 24394201 108 - 27905053 UAAACAAGAAAAGGCUGGGCAUGG-------AAUGGCGGAGAAUCAACGACGCUAAGACCCCGUCGUCGUUUAUCUCUGGCGUGGCUAUGUGAUAAAGUCGCGGUCCGUGUGGAA ..................((((((-------(..(.((((((...((((((((.........).)))))))..)))))).)((((((.........))))))..))))))).... ( -33.30, z-score = -0.70, R) >droSim1.chr3R 24085240 108 - 27517382 UAAACAAGAAAAGGCUGGGCAUGG-------AAUGGCGGAGAAUCAACGACGCUAAGACCCCGUCGUCGUUUAUCUCUGGCGUGGCUAUGUGAUAAAGUCGCGGUCCGCGUGGAA .............((.((((((((-------.(((.((((((...((((((((.........).)))))))..)))))).)))..))))(((((...))))).))))))...... ( -33.50, z-score = -0.41, R) >droSec1.super_22 997045 108 - 1066717 UAAACAAGAAAAGGCUGGGCAAGG-------AAUGGCGGAGAAUCAACGACGCUAAGACCCCGUCGUCGUUUAUCUCUGGCGUGGCUAUGUGAUAAAGUCGCGGUCCGCGUGGAA .............((.((((.((.-------.(((.((((((...((((((((.........).)))))))..)))))).)))..)).((((((...))))))))))))...... ( -32.20, z-score = -0.10, R) >droYak2.chr3R 6731054 115 + 28832112 UAAACAAGAAAAGGCUGGGCAUGGGGCAUGAAAUGGCGGAGAAUCAACGACGCUAAGACCCCGACGUCGUUUAUCUCUGGCGUGGCUAUGUGAUAAAGUCGCGGUCCGCGUGGAA ...................((((((((.......(.((((((...((((((((.........).)))))))..)))))).)((((((.........)))))).)))).))))... ( -34.00, z-score = 0.03, R) >droEre2.scaffold_4820 6831038 94 + 10470090 UAAACAAGUAAAGGCUGGGCAUGG-------AAUGGCGGAGAUUCAA--------------CGACGUCGUUUAUCUCUGGCGUGGCAAUGUGAUAAAGCCGCGGUCCGCGUGGAA .............((.((((....-------...(.(((((((..((--------------((....)))).))))))).)(((((...........))))).))))))...... ( -29.70, z-score = -0.96, R) >droAna3.scaffold_13340 17199495 83 - 23697760 ---------------------------UAAACAGGGCGGAGAAUCAACGCUUAAGA-----CCUCAGCGUUUAUCUCUGGCGUGGGCAAACGAUAAAGUCGCGGUACCAGUGGAU ---------------------------....((.(.((((((...((((((.....-----....))))))..)))))).).))..............((((.......)))).. ( -20.60, z-score = -0.10, R) >droWil1.scaffold_181089 9081962 87 - 12369635 ---------------------------UAAAUAAAGCAGAAAAUCAACGCUAAGACCCAGAGAGUGUGUGUUAUCUCUCUUGUGGC-AUAUGAUAAAGUCGCGGUCUGUGUAAAA ---------------------------...................((((..(((((.((((((..........)))))).(((((-..........)))))))))))))).... ( -20.40, z-score = -2.00, R) >consensus UAAACAAGAAAAGGCUGGGCAUGG_______AAUGGCGGAGAAUCAACGACGCUAAGACCCCGUCGUCGUUUAUCUCUGGCGUGGCUAUGUGAUAAAGUCGCGGUCCGCGUGGAA ................((((..............(.((((((...(((((((............)))))))..)))))).)(((((...........))))).))))........ (-16.95 = -18.87 + 1.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:37 2011