| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,372,177 – 24,372,270 |
| Length | 93 |
| Max. P | 0.977877 |

| Location | 24,372,177 – 24,372,270 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.24 |
| Shannon entropy | 0.41054 |
| G+C content | 0.50454 |
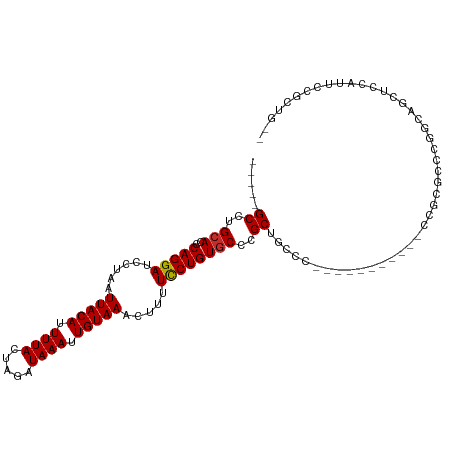
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -12.55 |
| Energy contribution | -12.41 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24372177 93 - 27905053 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUGCCC-----------CCGCGCCCUGCAGCUCCAUUCCGCUG-- -----((..((((..((((......(((((.((((.....)))).))))).....))))))))..(((((..-----------.........)))))........))..-- ( -19.90, z-score = -2.80, R) >droSim1.chr3R 24062760 93 - 27517382 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUUCCC-----------CCGCGCCCUGCAGCUCCAUUCCGCUG-- -----((..((((..((((......(((((.((((.....)))).))))).....))))))))..)).....-----------..........((((........))))-- ( -18.00, z-score = -2.66, R) >droSec1.super_22 975325 93 - 1066717 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUUCCC-----------CCGCGCCCUGCAGCUCCAUUCCGCUG-- -----((..((((..((((......(((((.((((.....)))).))))).....))))))))..)).....-----------..........((((........))))-- ( -18.00, z-score = -2.66, R) >droYak2.chr3R 6708258 104 + 28832112 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUGCCCCCGUCGCCCCGCCGCGCCCAGCAGCUCCAUUCCGCUG-- -----((..((((..((((......(((((.((((.....)))).))))).....))))))))..(((((...(((.((...)).)))....)))))........))..-- ( -24.50, z-score = -3.11, R) >droEre2.scaffold_4820 6808689 93 + 10470090 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUGCCC-----------CCGAGCCCUGCAGCUCCAUUCCGCUG-- -----((..((((..((((......(((((.((((.....)))).))))).....))))))))..(((((..-----------.........)))))........))..-- ( -19.90, z-score = -2.88, R) >droAna3.scaffold_13340 17176889 96 - 23697760 GGAGAGCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUUCUGUGCACGCGGCCCU-----CUACUCCGCCACUGGCCCACUUG---------- ((((.((((((((..((((......(((((.((((.....)))).))))).....)))))))))...))).))-----)).....(((...))).......---------- ( -22.10, z-score = -1.78, R) >dp4.chr2 15195151 95 + 30794189 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUUCUGUGCACGCUGCUG-----------CUGCCCCUGCCCCUGCCUCUGCUAUGGC -----(((.(((..((((.......(((((.((((.....)))).)))))...........(((.((....)-----------)))).......))))....)))...))) ( -19.40, z-score = -1.24, R) >droPer1.super_0 4814572 78 - 11822988 -----GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUUCUGUGCACGCUGCUG-----------CUGCCGCUGCC----------------- -----((.(((((..((((......(((((.((((.....)))).))))).....))))))))).)).((.(-----------(....)).)).----------------- ( -17.00, z-score = -1.74, R) >droWil1.scaffold_181089 9051957 84 - 12369635 -----GCCUGCA-CCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUUCUGUCCUCGCUGCUG----------CCUGCUGCUGUUGCUGCC----------- -----((..(((-.(((((......(((((.((((.....)))).))))).....))))).....((.((..----------...)).))...))).)).----------- ( -15.30, z-score = -0.89, R) >consensus _____GCCUGCACCCAGGAUCCUAAUUACAUUUUACUAGAUAAAUUGUAAACUUUUCCUGUGCCCGCUGCCC___________CCGCGCCCGGCAGCUCCAUUCCGCUG__ .....((..(((..(((((......(((((.((((.....)))).))))).....))))))))..))............................................ (-12.55 = -12.41 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:29 2011