| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,371,537 – 24,371,642 |
| Length | 105 |
| Max. P | 0.566147 |

| Location | 24,371,537 – 24,371,642 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.59 |
| Shannon entropy | 0.31340 |
| G+C content | 0.45626 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -24.27 |
| Energy contribution | -23.83 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24371537 105 - 27905053 UGUGUGUGCUGGCCCCUCACUAUGCAUGCGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG .((((((.(((.......(((((((((....)))))))))...((((((((((((.......))))))))))))....................))))))))).. ( -31.30, z-score = -0.71, R) >droSim1.chr3R 24062122 103 - 27517382 --UGUGUGCCGGCCCCUCACUAUGCAUGCGUGUGUGUAGUAUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG --.......((((.(.(((((((((((....)))))))))...((((((((((((.......))))))))))))......................)).).)))) ( -28.50, z-score = 0.02, R) >droSec1.super_22 974687 103 - 1066717 --UGUGUGCUGGCCCCUCACUAUGCAUGCGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCACUG --.((((((((.......(((((((((....)))))))))...((((((((((((.......))))))))))))....................))).))))).. ( -30.40, z-score = -1.03, R) >droEre2.scaffold_4820 6808063 99 + 10470090 ------UGCUGGCCCCUCACUAUGCAUGCGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG ------((((((.......))).))).((((((.(((.(((((((((((((((((.......)))))))))))))..............))))))).)))))).. ( -28.13, z-score = -0.76, R) >droAna3.scaffold_13340 17176297 97 - 23697760 --------GGGGAGGGGGGAUGAGCCUGCGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG --------...(((....((((((((((((....)))))....((((((((((((.......))))))))))))))).))))....))).....(((.....))) ( -28.10, z-score = -0.01, R) >droPer1.super_0 4813884 101 - 11822988 ----UCUCUCUUUCUCUCGAUGUGUGUGUGUGUGUGUAGUUUUGUGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG ----.................((((((.((((((((.(((...((((((((((((.......))))))))))))))).)))..........))))).)))))).. ( -26.40, z-score = -1.93, R) >droGri2.scaffold_14906 1411934 81 - 14172833 ------------------------UGUGUGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG ------------------------.((((((.(((((......((((((((((((.......))))))))))))(......).........))))).)))))).. ( -25.60, z-score = -1.94, R) >consensus ____U_UGCUGGCCCCUCACUAUGCAUGCGUGUGUGUAGUUUUGCGCAAAUAUUUGCAUCUGGAAUAUUUGCGCGCUCCAUCAUAAUUCAAACACAGAUGCGCUG ..........................(((((.(((((......((((((((((((.......))))))))))))(......).........))))).)))))... (-24.27 = -23.83 + -0.45)

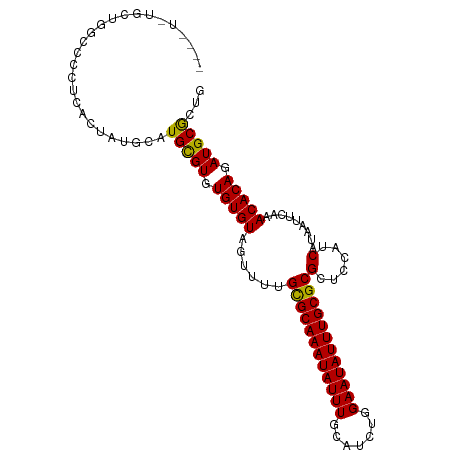

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:29 2011