| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,359,164 – 24,359,257 |
| Length | 93 |
| Max. P | 0.500000 |

| Location | 24,359,164 – 24,359,257 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.59439 |
| G+C content | 0.48915 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -12.19 |
| Energy contribution | -10.74 |
| Covariance contribution | -1.45 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
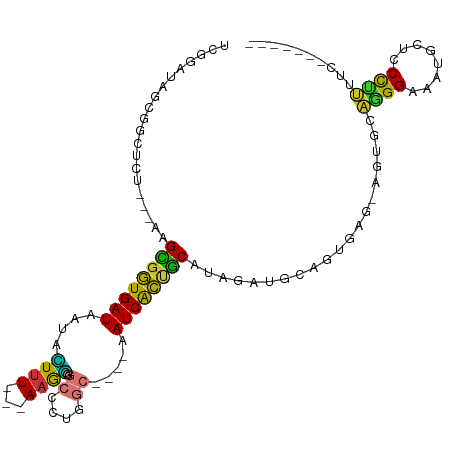
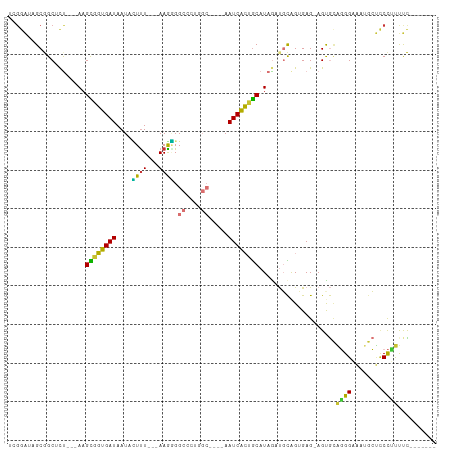
>dm3.chr3R 24359164 93 + 27905053 UCGGAUAGCGGCUCU---AAGCGGUGAUAAUACUCU---AAGGGGCCCUGGC----AAUCACUGCAUAGAUGCAGUGAG-AGUGCAGGGAAAUGCUCCCUUUUC------- .....(((.((((((---....((((....))))..---..)))))))))((----(.((((((((....)))))))).-..)))(((((.....)))))....------- ( -33.70, z-score = -1.80, R) >droSim1.chr3R 24049830 93 + 27517382 UCGGAUAGCGGCUCU---AAGCGGUGAUAAUACUUU---AAGGGGCCCUGGC----AAUCACUGCAUAGAUGCAGUGAG-AGUGCAGGGAAAUGCUCCCUUUUC------- .....(((.((((((---....((((....))))..---..)))))))))((----(.((((((((....)))))))).-..)))(((((.....)))))....------- ( -33.70, z-score = -2.03, R) >droSec1.super_22 962419 93 + 1066717 UCGGAUAGCGGCUCU---AAGCGGUGAUAAUACUUU---AAGGGGCCCUGGC----AAUCACUGCAUAGAUGCAGUGAG-AGUGCAGGGAAAUGCUCCCCUUUC------- .......((.((...---..)).))...........---(((((((((((.(----(.((((((((....)))))))).-..))))))).......))))))..------- ( -33.71, z-score = -1.78, R) >droYak2.chr3R 6693838 102 - 28832112 UCGGAUAGCGGCUCUC-UAAGCGGUGAUAAUAUUUUUUUAAGGGGCUCUGGCUGGCAAUCACUGCAUAAAUGCAGUGAG-AGUGGAGGGAAAUACUCCCUUUUC------- .(((.(((.(((((..-((((..(.........)..))))..)))))))).)))((..((((((((....)))))))).-.))(((((((.....)))))))..------- ( -32.30, z-score = -1.76, R) >droEre2.scaffold_4820 6793597 93 - 10470090 UCGGAUAGCGGCUCU---AAGCGGUGAUAAUAUUUU---AAGGGGCUCUGGC----AAUCACUGCAUAAAUGCAGUGAG-AGUGCAGGGAAAUGCUCCCUUUUC------- .......((.((...---..)).))...........---((((((((((.((----(.((((((((....)))))))).-..)))..)))...)).)))))...------- ( -30.10, z-score = -1.72, R) >droAna3.scaffold_13340 17163003 98 + 23697760 -----AAGCAGCGCU---AAGCGGUGAUAAUAUUUUUUUAAAAGGCAGCUGC----AAUCACUGCAUAGAUGCGGAGAG-AGCGCCGGGAAAGGCGGCCACCCUUUUACUU -----...((.(((.---..))).))............(((((((..(((((----..((.(((((....))))).)).-.))(((......))))))...)))))))... ( -28.70, z-score = -0.26, R) >dp4.chr2 20626832 93 - 30794189 -GGAGGAGCAGCAUCAAUAAGUGCUGAUAAUAGUUU---AAAUGGG----------AAUCGGCACAUAGACUUAGGGAGGAGCAUGCGGCGAAAGCUCUGCCCUUUU---- -(((((.(((((((......((((((((..((....---...))..----------.))))))))...(.(((......))))))))(((....))).)))))))).---- ( -26.70, z-score = -1.24, R) >droPer1.super_0 10314455 94 + 11822988 -GGAGGAGCAGCAUCAAUAAGUGCUGAUAAUAGUUU---AAAUGGG----------AAUCGGGGCAUAGACUUAGGGAGGAGCAUGCGGCGAAAGCUCUGCCCUUUUU--- -.(((...((((((......))))))......((((---..((...----------.))..)))).....)))(((((((.(((.(.(((....))))))))))))))--- ( -23.50, z-score = 0.14, R) >consensus UCGGAUAGCGGCUCU___AAGCGGUGAUAAUACUUU___AAGGGGCCCUGGC____AAUCACUGCAUAGAUGCAGUGAG_AGUGCAGGGAAAUGCUCCCUUUUC_______ ....................((((((((.............................))))))))....................((((........)))).......... (-12.19 = -10.74 + -1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:27 2011