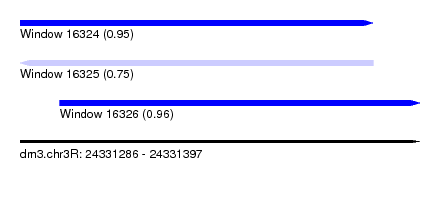
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,331,286 – 24,331,397 |
| Length | 111 |
| Max. P | 0.960343 |

| Location | 24,331,286 – 24,331,384 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.48522 |
| G+C content | 0.36083 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -13.98 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
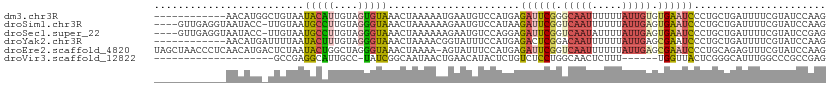
>dm3.chr3R 24331286 98 + 27905053 ------------GUUGUUA--CCUCAACAUGGC--UGUAAUACAUUGUAGUGUAAACUAAAAAUGAAUGUCCAUGAGAUUCGGGCAAUUUUUUAUUGUGUGAAUCCCUGCUGAU ------------.......--..(((.(((((.--....((.((((.((((....))))..)))).))..))))).((((((.(((((.....))))).)))))).....))). ( -21.10, z-score = -1.14, R) >droSim1.chr3R 24021117 105 + 27517382 ---------GUUGUUACCAUGUUGAGGUAAUACCUUGUAAUGCCUUGUAGGGUAAACUAAAAAAGAAUGUCCAUAAGAUUCGGUCAAUUUUUUAUUGAGUGAAUCCCUGCUGAU ---------....(((((....(((((((.(((...))).)))))))...))))).....................((((((.(((((.....))))).))))))......... ( -23.90, z-score = -1.93, R) >droSec1.super_22 934403 105 + 1066717 ---------GUUGUUACCAUGUUGAGGUAAUACCUUGUAAUGCCUUGUAGGGUAAACUAAAAAAGAAUGUCCAGGAGAUUCGGUCAAUAUUUUAUUGAGUGAAUCCCUGCUGAU ---------....(((((....(((((((.(((...))).)))))))...)))))................((((.((((((.((((((...)))))).))))))))))..... ( -30.50, z-score = -3.46, R) >droYak2.chr3R 6665660 87 - 28832112 -------------------------AACAUGAU--UUUAAUACUUUGUAGGGUAAACUAAAACGGUAUUUCCAUGAGACUCGGACAAUUUUUUAUUGAGCGAAUCCCUGCUGAU -------------------------........--...........((((((...........((.....)).......(((..((((.....))))..)))..)))))).... ( -14.10, z-score = -0.17, R) >droEre2.scaffold_4820 6763072 111 - 10470090 GUUGUUAUAUUCCUAGCUAACCCUCAACAUGAC--UCUAAUACUGGCUAGGGUAAACUAAAA-AGUAUUUCCAUGAGAUUCGGUCAAUUUUUUAUUGAGCGAAUCCCUGCAGAG (((((((......)))).))).(((..((((..--...((((((...(((......)))...-))))))..)))).((((((.(((((.....))))).))))))......))) ( -25.40, z-score = -1.83, R) >consensus __________U_GUUACCA__CCGAAACAUGAC__UGUAAUACCUUGUAGGGUAAACUAAAAAAGAAUGUCCAUGAGAUUCGGUCAAUUUUUUAUUGAGUGAAUCCCUGCUGAU ........................................(((((....)))))......................((((((.(((((.....))))).))))))......... (-13.98 = -13.38 + -0.60)

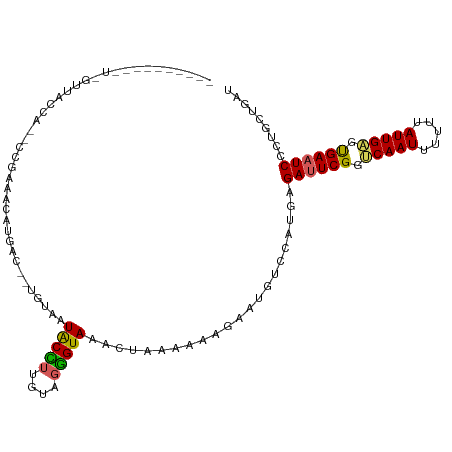
| Location | 24,331,286 – 24,331,384 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.63 |
| Shannon entropy | 0.48522 |
| G+C content | 0.36083 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -10.16 |
| Energy contribution | -10.00 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24331286 98 - 27905053 AUCAGCAGGGAUUCACACAAUAAAAAAUUGCCCGAAUCUCAUGGACAUUCAUUUUUAGUUUACACUACAAUGUAUUACA--GCCAUGUUGAGG--UAACAAC------------ .(((((.(((((((...((((.....))))...)))))))((((.....((((..((((....)))).)))).......--.)))))))))..--.......------------ ( -19.62, z-score = -1.88, R) >droSim1.chr3R 24021117 105 - 27517382 AUCAGCAGGGAUUCACUCAAUAAAAAAUUGACCGAAUCUUAUGGACAUUCUUUUUUAGUUUACCCUACAAGGCAUUACAAGGUAUUACCUCAACAUGGUAACAAC--------- ....((.(((((((..(((((.....)))))..))))))).(((((...........))))).........))...........(((((.......)))))....--------- ( -15.90, z-score = -0.36, R) >droSec1.super_22 934403 105 - 1066717 AUCAGCAGGGAUUCACUCAAUAAAAUAUUGACCGAAUCUCCUGGACAUUCUUUUUUAGUUUACCCUACAAGGCAUUACAAGGUAUUACCUCAACAUGGUAACAAC--------- (((..(((((((((..((((((...))))))..))))).))))..............((....((.....))....))..))).(((((.......)))))....--------- ( -20.50, z-score = -1.67, R) >droYak2.chr3R 6665660 87 + 28832112 AUCAGCAGGGAUUCGCUCAAUAAAAAAUUGUCCGAGUCUCAUGGAAAUACCGUUUUAGUUUACCCUACAAAGUAUUAAA--AUCAUGUU------------------------- ...((((((((((((..((((.....))))..)))))))).....(((((..((.(((......))).)).)))))...--....))))------------------------- ( -16.30, z-score = -1.78, R) >droEre2.scaffold_4820 6763072 111 + 10470090 CUCUGCAGGGAUUCGCUCAAUAAAAAAUUGACCGAAUCUCAUGGAAAUACU-UUUUAGUUUACCCUAGCCAGUAUUAGA--GUCAUGUUGAGGGUUAGCUAGGAAUAUAACAAC .......((((((((.(((((.....))))).)))))))).((...(((.(-((((((((.(((((...((........--....))...))))).))))))))))))..)).. ( -29.30, z-score = -2.36, R) >consensus AUCAGCAGGGAUUCACUCAAUAAAAAAUUGACCGAAUCUCAUGGACAUUCUUUUUUAGUUUACCCUACAAAGUAUUACA__GUCAUGUUGAAA__UAGCAAC_A__________ ....((.(((((((..(((((.....)))))..)))))))...............(((......)))....))......................................... (-10.16 = -10.00 + -0.16)

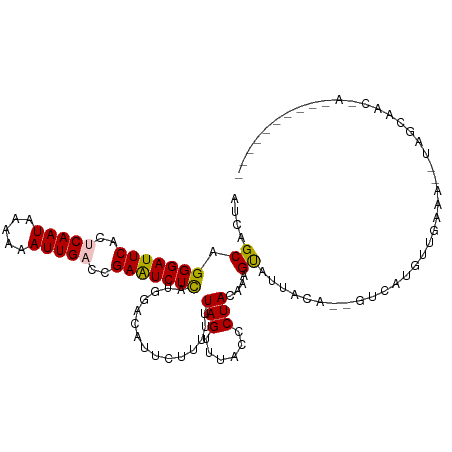
| Location | 24,331,297 – 24,331,397 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 65.47 |
| Shannon entropy | 0.64849 |
| G+C content | 0.39448 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -9.95 |
| Energy contribution | -10.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24331297 100 + 27905053 ------------AACAUGGCUGUAAUACAUUGUAGUGUAAACUAAAAAUGAAUGUCCAUGAGAUUCGGGCAAUUUUUUAUUGUGUGAAUCCCUGCUGAUUUUCGUAUCCAAG ------------....(((......(((((....)))))........(((((.(((((.(.((((((.(((((.....))))).))))))).))..))).)))))..))).. ( -21.30, z-score = -1.32, R) >droSim1.chr3R 24021128 107 + 27517382 ----GUUGAGGUAAUACC-UUGUAAUGCCUUGUAGGGUAAACUAAAAAAGAAUGUCCAUAAGAUUCGGUCAAUUUUUUAUUGAGUGAAUCCCUGCUGAUUUUCGUAUCCAAG ----...((((((.(((.-..))).))))))...(((((..........(((.(((.....((((((.(((((.....))))).))))))......))).))).)))))... ( -24.60, z-score = -2.16, R) >droSec1.super_22 934414 107 + 1066717 ----GUUGAGGUAAUACC-UUGUAAUGCCUUGUAGGGUAAACUAAAAAAGAAUGUCCAGGAGAUUCGGUCAAUAUUUUAUUGAGUGAAUCCCUGCUGAUUUUCGUAUCCGAG ----...((((((.(((.-..))).))))))...(((((..........(((.(((((((.((((((.((((((...)))))).))))))))))..))).))).)))))... ( -31.80, z-score = -3.71, R) >droYak2.chr3R 6665660 100 - 28832112 ------------AACAUGAUUUUAAUACUUUGUAGGGUAAACUAAAACGGUAUUUCCAUGAGACUCGGACAAUUUUUUAUUGAGCGAAUCCCUGCUGAUUUUCGUAUCCAAG ------------..((((.....((((((...(((......)))....))))))..))))......(((((((.....)))).(((((............))))).)))... ( -15.60, z-score = 0.32, R) >droEre2.scaffold_4820 6763085 111 - 10470090 UAGCUAACCCUCAACAUGACUCUAAUACUGGCUAGGGUAAACUAAAA-AGUAUUUCCAUGAGAUUCGGUCAAUUUUUUAUUGAGCGAAUCCCUGCAGAGUUUCGUAUCCAAG .............((..((((((((((((...(((......)))...-))))))..((.(.((((((.(((((.....))))).))))))).)).))))))..))....... ( -25.80, z-score = -1.94, R) >droVir3.scaffold_12822 3771643 85 + 4096053 --------------------GCCGAGGCAUUGCC-UAUCGGCAAUAACUGAACAUACUCUGUCUCCUGGCAACUCUUU------UGGUUACUCGGGCAUUUGGCCCGCCGAG --------------------((((((((((((((-....))))))....((......)).)))))..))).......(------(((.....(((((.....))))))))). ( -27.70, z-score = -2.21, R) >consensus ____________AACAUG_UUGUAAUACAUUGUAGGGUAAACUAAAAAAGAAUGUCCAUGAGAUUCGGUCAAUUUUUUAUUGAGUGAAUCCCUGCUGAUUUUCGUAUCCAAG .........................(((((....)))))......................((((((.(((((.....))))).))))))...................... ( -9.95 = -10.15 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:23 2011