| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,316,273 – 24,316,396 |
| Length | 123 |
| Max. P | 0.938705 |

| Location | 24,316,273 – 24,316,371 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.38870 |
| G+C content | 0.38294 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.25 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
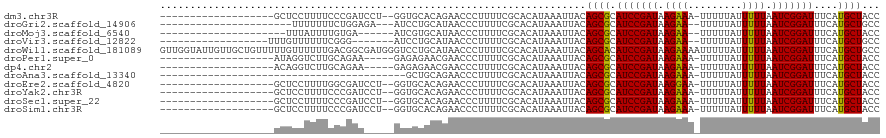
>dm3.chr3R 24316273 98 + 27905053 -------------------GCUCCUUUUCCCGAUCCU--GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...........(((....--(((.......)))....)))............((((.(((((((.(((((-(....)))))).)))))))....))))... ( -17.90, z-score = -1.91, R) >droGri2.scaffold_14906 1353567 93 + 14172833 ----------------------UUUUUUUCUGGAGA---AUCCUGCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA--UUUUUAUUUUUAAUCGGAUUUCAUGCUGCC ----------------------.........((...---..))(((............))).........(((((.((((((((((((--(....)))))).)))))))....))))).. ( -16.80, z-score = -1.52, R) >droMoj3.scaffold_6540 17069359 91 + 34148556 ---------------------UUUAUUUUGUGA------AUCGUGCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA--UUUUUAUUUUUAAUCGGAUUUCAUGCUACC ---------------------(((((..(((((------(..((.....))....)))))).)))))....((((.((((((((((((--(....)))))).)))))))....))))... ( -17.80, z-score = -2.62, R) >droVir3.scaffold_12822 3755887 93 + 4096053 ------------------UUUGUUUUUUCGGG-------AUCCUGCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA--UUUUUAUUUUUAAUCGGAUUUCAUGCUGCC ------------------...........(((-------...........))).................(((((.((((((((((((--(....)))))).)))))))....))))).. ( -18.00, z-score = -1.54, R) >droWil1.scaffold_181089 8992921 120 + 12369635 GUUGGUAUUGUUGCUGUUUUUGUUUUUUGACGGCGAUGGGUCCUGCAUAACCCUUUUCGCACAUAAAUUACAGCACAUCCGAUAAGAAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUGCC ...(((((...((((((..(((((....))).((((.((((........))))...)))).....))..)))))).(((((((.(((((.......))))).)))))))...)))))... ( -31.20, z-score = -2.48, R) >droPer1.super_0 10269999 95 + 11822988 -------------------AUAGGUCUUGCAGAA-----GAGAGAACGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...(((..(((.(((-----(((.(......).)))))))))..........((((.(((((((.(((((-(....)))))).)))))))....))))))) ( -21.60, z-score = -2.90, R) >dp4.chr2 20582403 95 - 30794189 -------------------ACAGGUCUUGCAGAA-----GAGAGAACGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...(((..(((.(((-----(((.(......).)))))))))..........((((.(((((((.(((((-(....)))))).)))))))....))))))) ( -21.60, z-score = -3.05, R) >droAna3.scaffold_13340 17120220 78 + 23697760 -----------------------------------------GCUGCAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -----------------------------------------...((((((.......(((............))).(((((((.(((((-(....)))))).)))))))))).))).... ( -15.90, z-score = -2.99, R) >droEre2.scaffold_4820 6748049 98 - 10470090 -------------------GCUCCUUUUGGCGAUCCU--GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGGAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------(((.....((((((....--(((.......)))....)))).))........)))(((((((((.(((((-(....)))))).)))))).....))).... ( -22.22, z-score = -1.96, R) >droYak2.chr3R 6650486 98 - 28832112 -------------------GCUCCUUUUCCCGAUCCU--GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...........(((....--(((.......)))....)))............((((.(((((((.(((((-(....)))))).)))))))....))))... ( -17.90, z-score = -1.91, R) >droSec1.super_22 919325 98 + 1066717 -------------------GCUCCUUUUCCCGAUCCU--GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...........(((....--(((.......)))....)))............((((.(((((((.(((((-(....)))))).)))))))....))))... ( -17.90, z-score = -1.91, R) >droSim1.chr3R 24008132 98 + 27517382 -------------------GCUCCUUUUCCCGAUCCU--GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACC -------------------...........(((....--(((.......)))....)))............((((.(((((((.(((((-(....)))))).)))))))....))))... ( -17.90, z-score = -1.91, R) >consensus ___________________GCUCCUUUUCCCGAU_____GGUGCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAA_UUUUUAUUUUUAAUCGGAUUUCAUGCUACC .......................................................................((((.((((((((((((........))))).)))))))....))))... (-11.40 = -11.25 + -0.15)
| Location | 24,316,294 – 24,316,396 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Shannon entropy | 0.29579 |
| G+C content | 0.42886 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -12.95 |
| Energy contribution | -12.81 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24316294 102 + 27905053 GCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCGU--ACUCCGUUGUC-------- (((..(((.......(((............))).(((((((.((((((....)))))).)))))))))).)))......((((((.(....--.).))))))..-------- ( -21.80, z-score = -2.47, R) >droGri2.scaffold_14906 1353586 101 + 14172833 --GCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUGCCAUCAACGGCGCCAGCGCCAGCGUCGAU-------- --.............(((..........(((((.((((((((((((-(....)))))).)))))))....)))))........((((....)))).))).....-------- ( -27.10, z-score = -2.83, R) >droMoj3.scaffold_6540 17069376 102 + 34148556 --GCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCAGCUUCAGCGUAGACA------- --........((...(((...........((((.((((((((((((-(....)))))).)))))))....)))).........(((....)))...))).))...------- ( -19.50, z-score = -1.75, R) >droVir3.scaffold_12822 3755906 101 + 4096053 --GCAUAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAA-UUUUUAUUUUUAAUCGGAUUUCAUGCUGCCAUCAACGGCGCCAGCGUCAGCGUCAGC-------- --.............(((..........(((((.((((((((((((-(....)))))).)))))))....)))))........((((....)))).))).....-------- ( -24.40, z-score = -2.23, R) >droPer1.super_0 10270020 108 + 11822988 --ACG-AACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGUGGCGCCACCAUCUCGCGCCUUUGUC- --.((-((.....)))).............(((((((((((.((((((....)))))).))))))).....((((((......))))))...........)))).......- ( -29.50, z-score = -4.10, R) >dp4.chr2 20582424 108 - 30794189 --ACG-AACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGUGGCGCCACCAUCUCGCGCCUUUGUC- --.((-((.....)))).............(((((((((((.((((((....)))))).))))))).....((((((......))))))...........)))).......- ( -29.50, z-score = -4.10, R) >droAna3.scaffold_13340 17120223 110 + 23697760 --GCAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCACCAUCGCCGUUGUGUCCUUGUC --((((((.......(((............))).(((((((.((((((....)))))).)))))))))).)))...((.((((((((.......))))))))))........ ( -28.60, z-score = -3.80, R) >droEre2.scaffold_4820 6748070 102 - 10470090 GCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGGAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGACGCCGU--UCUCCGUUGUC-------- (((..(((.......(((............))).(((((((.((((((....)))))).)))))))))).)))........((((((....--....)))))).-------- ( -18.80, z-score = -1.42, R) >droYak2.chr3R 6650507 102 - 28832112 GCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCGU--UCUCCGUUGUC-------- (((..(((.......(((............))).(((((((.((((((....)))))).)))))))))).)))......((((((.(....--.).))))))..-------- ( -21.20, z-score = -2.19, R) >droSec1.super_22 919346 102 + 1066717 GCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCGU--ACUCCGUUGUC-------- (((..(((.......(((............))).(((((((.((((((....)))))).)))))))))).)))......((((((.(....--.).))))))..-------- ( -21.80, z-score = -2.47, R) >droSim1.chr3R 24008153 102 + 27517382 GCACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCGU--ACUCCGUUGUC-------- (((..(((.......(((............))).(((((((.((((((....)))))).)))))))))).)))......((((((.(....--.).))))))..-------- ( -21.80, z-score = -2.47, R) >consensus __ACAGAACCCUUUUCGCACAUAAAUUACAGCGCAUCCGAUAAGAAAUUUUUAUUUUUAAUCGGAUUUCAUGCUACCAUCAACGGCGCCGUC_UCACCGUUGUC________ ...............(((...........((((.(((((((.((((........)))).)))))))....))))..........)))......................... (-12.95 = -12.81 + -0.14)

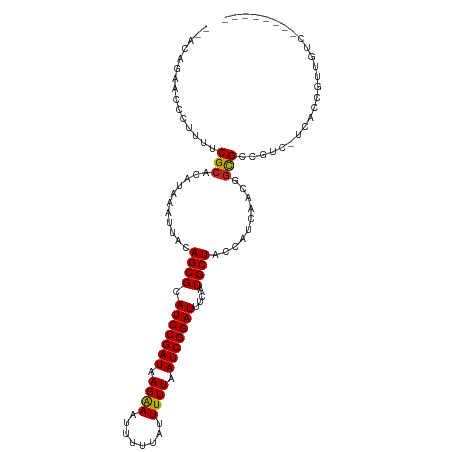
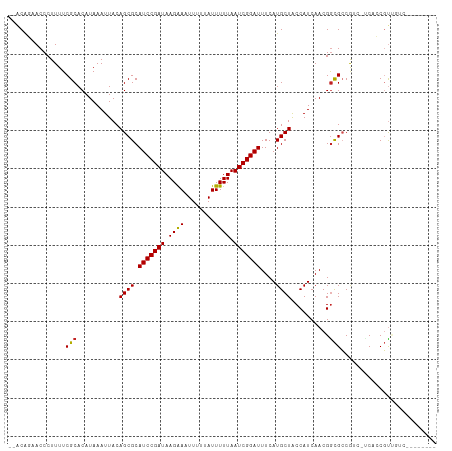
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:19 2011