
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,293,644 – 24,293,762 |
| Length | 118 |
| Max. P | 0.791205 |

| Location | 24,293,644 – 24,293,762 |
|---|---|
| Length | 118 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.95 |
| Shannon entropy | 0.54914 |
| G+C content | 0.46061 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
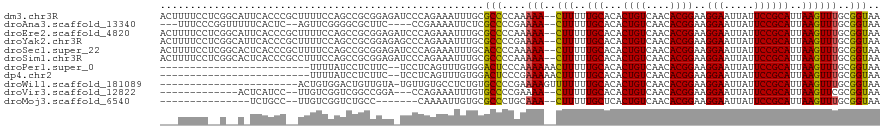
>dm3.chr3R 24293644 118 + 27905053 ACUUUUCCUCGGCAUUCACCCGCUUUUCCAGCCGCGGAGAUCCCAGAAAUUUGCGCCCCAAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ...(((((.((((.................)))).)))))..............(((....(((--(((..(((...((((....))))..(((.....))))))..))))))..))).. ( -28.33, z-score = -1.06, R) >droAna3.scaffold_13340 17093147 109 + 23697760 ---UUUCCCGGUUUUUCACUC--AGUUCGGGGCGCUUC----CCGAAAAUUCUCGCCCCGAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ---....(((...........--...((((((((....----...........))))))))(((--(((..(((...((((....))))..(((.....))))))..)))))).)))... ( -31.76, z-score = -2.09, R) >droEre2.scaffold_4820 6725146 118 - 10470090 ACUUUUCCUCGGCAUUCACCCGCUUUUCCAGCCGCGGAGAUCCCAGAAAUUUGCGCCCCAAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ...(((((.((((.................)))).)))))..............(((....(((--(((..(((...((((....))))..(((.....))))))..))))))..))).. ( -28.33, z-score = -1.06, R) >droYak2.chr3R 6627653 118 - 28832112 ACUUUUCCUCGGCAUUCACCCGCUUUUCCAGCCGCGGAGAGCCCAGAAAUUUGCGCCCCGAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ...(((((.((((.................)))).)))))((.(((....))).)).(((.(((--(((..(((...((((....))))..(((.....))))))..)))))).)))... ( -31.63, z-score = -1.41, R) >droSec1.super_22 898055 118 + 1066717 ACUUUUCCUCGGCACUCACCCGCUUUUCCAGCCGCGGAGAUCCCAGAAAUUUGCACCCCAAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ...(((((.((((.................)))).)))))..............(((....(((--(((..(((...((((....))))..(((.....))))))..))))))..))).. ( -28.03, z-score = -1.48, R) >droSim1.chr3R 23986781 118 + 27517382 ACUUUUCCUCGGCACUCACCCGCCUUUCCAGCCGCGGAGAUCCCAGAAAUUUGCGCCCCAAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ...(((((.((((.................)))).)))))..............(((....(((--(((..(((...((((....))))..(((.....))))))..))))))..))).. ( -28.33, z-score = -1.03, R) >droPer1.super_0 10242909 93 + 11822988 -------------------------UUUUAUCCUCUUC--UCCUCAGUUUGUGGACUCCCAAAAAACUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA -------------------------.....((((((..--....((((.((..((..............))..)).)))).......)).))))......(((((.......)))))... ( -18.26, z-score = -0.96, R) >dp4.chr2 20555836 93 - 30794189 -------------------------UUUUAUCCUCUUC--UCCUCAGUUUGUGGACUCCCGAAAAACUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA -------------------------.............--(((.(.....).)))...(((..((((((..(((...((((....))))..(((.....))))))..)))))).)))... ( -19.60, z-score = -1.11, R) >droWil1.scaffold_181089 8965632 96 + 12369635 -----------------------ACUGUGGACUGUUGUA-UGUUGUGCCUCUGUGCCCCGAAAAGUUUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA -----------------------((((..((((..(((.-(((((.((...(((((...((((....)))).)))))..))))))).((((.......)))))))...))))..)))).. ( -26.80, z-score = -1.41, R) >droVir3.scaffold_12822 3730966 100 + 4096053 -------------ACUCAUCC--UUGUCGGUCGGCCGGA---CCAGAAAUUUGUGCCCCGAAAA--CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUCGCGGUAA -------------........--.....((((.....))---)).........((((.(((..(--(((..(((...((((....))))..(((.....))))))..))))))).)))). ( -25.10, z-score = -0.29, R) >droMoj3.scaffold_6540 17042710 94 + 34148556 ---------------UCUGCC--UUGUCGGUCUGCC-------CAAAAUUGUGCGCCCUGCAAA--CUUUUUGCUCACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ---------------..((((--.....(((..((.-------((....)).)))))..(((((--(((..(((...((((....))))..(((.....))))))..)))))))))))). ( -27.30, z-score = -2.07, R) >consensus ___UUUCC__G__A_UCACCC__UUUUCCAGCCGCGGAG_UCCCAGAAAUUUGCGCCCCGAAAA__CUUUUUGCACACUGUCAACACGGAAGGAAUUAUUCCGCAUUAAGUUUGCGGUAA ......................................................(((.((((....(((..(((...((((....))))..(((.....))))))..))))))).))).. (-13.84 = -13.30 + -0.54)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:18 2011