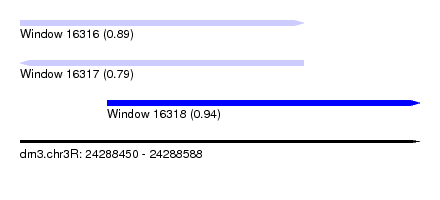
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,288,450 – 24,288,588 |
| Length | 138 |
| Max. P | 0.937390 |

| Location | 24,288,450 – 24,288,548 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.46 |
| Shannon entropy | 0.14736 |
| G+C content | 0.35034 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -16.81 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
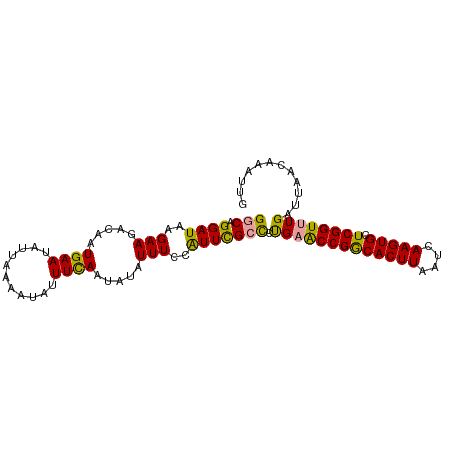
>dm3.chr3R 24288450 98 + 27905053 GUCAGGAUAAGAAGACAAUGAAUAUUAAAACAUUUUAAUAUAUUUUCAUUCGCUCUGAGCCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUG .(((((....(((((.(((..((((((((....))))))))))).)).)))..)))))((((((((((....)))))).))))............... ( -21.70, z-score = -2.49, R) >droYak2.chr3R 6622269 98 - 28832112 GGCAGGAUAAGAAGGCCAUGAAUAUUAAAAUAUUUCAAUAUAUUUCCAUUUGCCCCGAACCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUAG ((((((....((((....((((...........)))).....))))..)))))).(((((((((((((....)))))).)))))))............ ( -25.40, z-score = -2.97, R) >droEre2.scaffold_4820 6719799 98 - 10470090 GGCAGGAUAAGAAGCGAAUGAAUAUUAAAAUAUUUCAAUAUAUUUCCGUUCGCCCUGUACCGACACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUG .(((((.......((((((((((((..............)))))..))))))))))))((((((((((....))))).)))))............... ( -23.45, z-score = -2.71, R) >consensus GGCAGGAUAAGAAGACAAUGAAUAUUAAAAUAUUUCAAUAUAUUUCCAUUCGCCCUGAACCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUG (((.((((..(((.....((((...........)))).....)))..))))))).(((((((((((((....))))).))))))))............ (-16.81 = -15.93 + -0.87)

| Location | 24,288,450 – 24,288,548 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.46 |
| Shannon entropy | 0.14736 |
| G+C content | 0.35034 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.47 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24288450 98 - 27905053 CAAUUUGUUAAUCAAACCGAGCACUUGAUUAAGUGCCGGCUCAGAGCGAAUGAAAAUAUAUUAAAAUGUUUUAAUAUUCAUUGUCUUCUUAUCCUGAC ..(((((((.......(((.((((((....))))))))).....)))))))((....((((((((....))))))))......))............. ( -18.20, z-score = -1.45, R) >droYak2.chr3R 6622269 98 + 28832112 CUAUUUGUUAAUCAAACCGAGCACUUGAUUAAGUGCCGGUUCGGGGCAAAUGGAAAUAUAUUGAAAUAUUUUAAUAUUCAUGGCCUUCUUAUCCUGCC ((((((((...((.(((((.((((((....))))))))))).)).))))))))....((((((((....))))))))....(((...........))) ( -25.70, z-score = -2.37, R) >droEre2.scaffold_4820 6719799 98 + 10470090 CAAUUUGUUAAUCAAACCGAGCACUUGAUUAAGUGUCGGUACAGGGCGAACGGAAAUAUAUUGAAAUAUUUUAAUAUUCAUUCGCUUCUUAUCCUGCC ...............(((((.(((((....)))))))))).(((((.(((((((...((((((((....))))))))...)))).)))...))))).. ( -24.10, z-score = -2.69, R) >consensus CAAUUUGUUAAUCAAACCGAGCACUUGAUUAAGUGCCGGUUCAGGGCGAAUGGAAAUAUAUUGAAAUAUUUUAAUAUUCAUUGCCUUCUUAUCCUGCC ..(((((((.....(((((.((((((....)))))))))))...))))))).(((((((......))))))).......................... (-17.57 = -17.47 + -0.10)

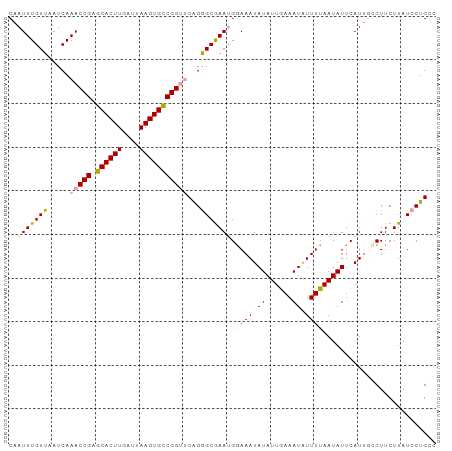
| Location | 24,288,480 – 24,288,588 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Shannon entropy | 0.19556 |
| G+C content | 0.39506 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.20 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
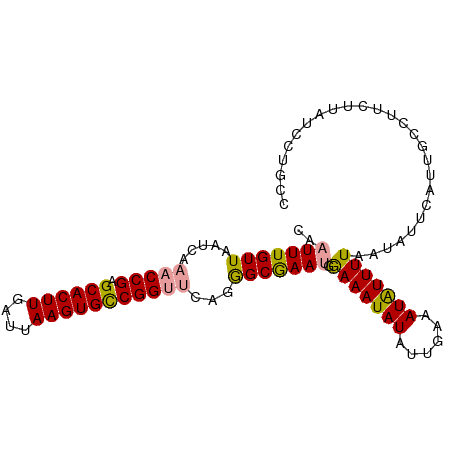
>dm3.chr3R 24288480 108 + 27905053 CAUUUUAAUAUAUUUUCAUUCGCUCUGAGCCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUGUGUUUCCCGCGAUCUCGAAAUAUACAUUUAGGUCCGAACU ........((((((((...((((...((((((((((((....)))))).))))))((...((.....))...))..))))....))))))))................ ( -25.30, z-score = -2.35, R) >droYak2.chr3R 6622299 108 - 28832112 UAUUUCAAUAUAUUUCCAUUUGCCCCGAACCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUAGGGCUGCCCGCGGUCAGGAAAUAUUCAUUUAAGACCGAAAU .(((((...(((((((((((((...(((((((((((((....)))))).))))))).....)))))..(((((....))))).))))))))((......))..))))) ( -33.80, z-score = -3.83, R) >droEre2.scaffold_4820 6719829 108 - 10470090 UAUUUCAAUAUAUUUCCGUUCGCCCUGUACCGACACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUGGGUUGCCCGCGGUCACGAAAUAUUUAUUCAAGUCCGAAAU .........(((((((...((((.....((((((((((....))))).)))))..............(((...)))))))....)))))))................. ( -21.70, z-score = -0.62, R) >consensus UAUUUCAAUAUAUUUCCAUUCGCCCUGAACCGGCACUUAAUCAAGUGCUCGGUUUGAUUAACAAAUUGGGUUGCCCGCGGUCACGAAAUAUUCAUUUAAGUCCGAAAU .........(((((((...((((..(((((((((((((....))))).))))))))...........(((...)))))))....)))))))................. (-21.85 = -21.20 + -0.65)

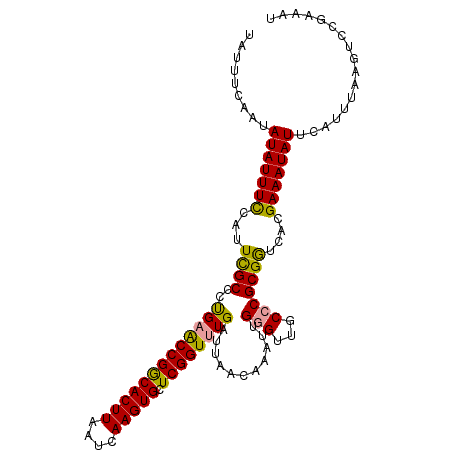
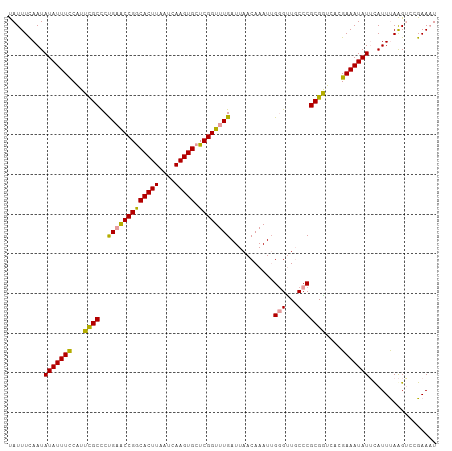
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:17 2011