
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,886,357 – 8,886,449 |
| Length | 92 |
| Max. P | 0.857622 |

| Location | 8,886,357 – 8,886,449 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.31615 |
| G+C content | 0.40161 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -13.96 |
| Energy contribution | -14.96 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
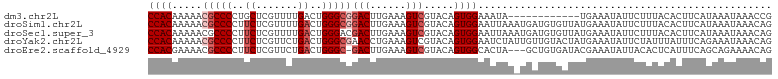
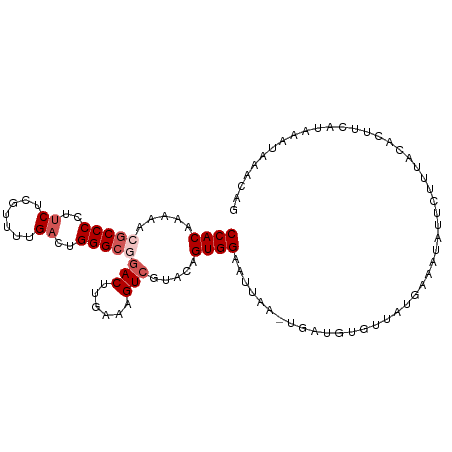
>dm3.chr2L 8886357 92 + 23011544 CCACAAAAACGCCCCUGCUCGUUUUGACUGGGCGGACUUGAAAGUCGUACAGUGGAAAUA------------UGAAAUAUUCUUUACACUUCAUAAAUAAACCG ((((.....(((((...(.......)...)))))((((....)))).....))))...((------------((((.............))))))......... ( -20.62, z-score = -2.07, R) >droSim1.chr2L 8665378 104 + 22036055 CCACAAAAACGCCCCUUCUCGUUUUGACUGGGCGGACUUGAAAGUCGUACAGUGGAAUUAAAUGAUGUGUUAUGAAAUAUUCUUUACACUUCAUAAAUAAACAG ((((.....(((((..((.......))..)))))((((....)))).....))))......((((.((((...((.....))...)))).)))).......... ( -27.30, z-score = -3.28, R) >droSec1.super_3 4360276 104 + 7220098 CCACAAAAACGCCCCUUCUCGUUUUGACUGGGACGACUUGAAAGUCGUACAGUGGAAUUAAAUGAUGUGUUAUGAAAUAUUCUUUACACUUCAUAAAUAAACAG ((((.......(((..((.......))..)))((((((....))))))...))))......((((.((((...((.....))...)))).)))).......... ( -24.50, z-score = -2.65, R) >droYak2.chr2L 11534230 104 + 22324452 CCACAAAAACGCCCCUUCUCGUUCUGACUGGGCGAACCUGAAAGUCGUACAGUGGAAUCUAUUGUUGUACUAUGAAAUAUUCUAUUUAUUUCAGAAAUAAACAG .........(((((..((.......))..)))))...(((......((((((..(......)..))))))..(((((((.......)))))))........))) ( -23.50, z-score = -2.44, R) >droEre2.scaffold_4929 9479659 100 + 26641161 CCACGAAAACGCCCCUUCUCGUUCUGACUGGGC-GACUUGAAAGUCGUACAGUGGCACUA---GCUGUGAUACGAAAUAUUACACUCAUUUCAGCAGAAAACAG ....((((.(((((..((.......))..))))-).........(((((..(..((....---))..)..))))).............))))............ ( -22.60, z-score = -0.72, R) >consensus CCACAAAAACGCCCCUUCUCGUUUUGACUGGGCGGACUUGAAAGUCGUACAGUGGAAUUAA_UGAUGUGUUAUGAAAUAUUCUUUACACUUCAUAAAUAAACAG ((((.....(((((..((.......))..)))))(((......))).....))))................................................. (-13.96 = -14.96 + 1.00)
| Location | 8,886,357 – 8,886,449 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 82.55 |
| Shannon entropy | 0.31615 |
| G+C content | 0.40161 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.12 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
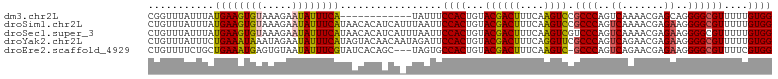
>dm3.chr2L 8886357 92 - 23011544 CGGUUUAUUUAUGAAGUGUAAAGAAUAUUUCA------------UAUUUCCACUGUACGACUUUCAAGUCCGCCCAGUCAAAACGAGCAGGGGCGUUUUUGUGG .........((((((((((.....))))))))------------))...((((.....((((....))))(((((..((.....))....))))).....)))) ( -23.20, z-score = -1.83, R) >droSim1.chr2L 8665378 104 - 22036055 CUGUUUAUUUAUGAAGUGUAAAGAAUAUUUCAUAACACAUCAUUUAAUUCCACUGUACGACUUUCAAGUCCGCCCAGUCAAAACGAGAAGGGGCGUUUUUGUGG ........(((((((((((.....)))))))))))..............((((.....((((....))))(((((..((.......))..))))).....)))) ( -26.50, z-score = -2.66, R) >droSec1.super_3 4360276 104 - 7220098 CUGUUUAUUUAUGAAGUGUAAAGAAUAUUUCAUAACACAUCAUUUAAUUCCACUGUACGACUUUCAAGUCGUCCCAGUCAAAACGAGAAGGGGCGUUUUUGUGG ........(((((((((((.....)))))))))))..............((((...((((((....))))(((((..((.......)).)))))))....)))) ( -25.10, z-score = -2.26, R) >droYak2.chr2L 11534230 104 - 22324452 CUGUUUAUUUCUGAAAUAAAUAGAAUAUUUCAUAGUACAACAAUAGAUUCCACUGUACGACUUUCAGGUUCGCCCAGUCAGAACGAGAAGGGGCGUUUUUGUGG (((((((((.....)))))))))...........(((((..............)))))......((((..(((((..((.......))..)))))..))))... ( -23.24, z-score = -1.15, R) >droEre2.scaffold_4929 9479659 100 - 26641161 CUGUUUUCUGCUGAAAUGAGUGUAAUAUUUCGUAUCACAGC---UAGUGCCACUGUACGACUUUCAAGUC-GCCCAGUCAGAACGAGAAGGGGCGUUUUCGUGG .......(((.(((.(((((........))))).)))))).---.....((((...((((((....))))-((((..((.......))..))))))....)))) ( -25.50, z-score = -0.26, R) >consensus CUGUUUAUUUAUGAAGUGUAAAGAAUAUUUCAUAACACAUCA_UUAAUUCCACUGUACGACUUUCAAGUCCGCCCAGUCAAAACGAGAAGGGGCGUUUUUGUGG ...........((((((((.....)))))))).................((((...((((((....)))).((((..((.......))..))))))....)))) (-19.20 = -19.12 + -0.08)

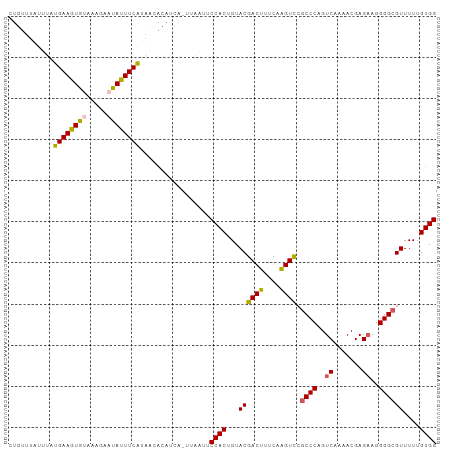
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:15 2011