
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,284,462 – 24,284,565 |
| Length | 103 |
| Max. P | 0.617631 |

| Location | 24,284,462 – 24,284,565 |
|---|---|
| Length | 103 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.23 |
| Shannon entropy | 0.58893 |
| G+C content | 0.51416 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.27 |
| Covariance contribution | -0.85 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.617631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24284462 103 - 27905053 --AUCGCAUUAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUUCUCAUUAGGUUUAGUGCC-CCGUGCGAG---GCGACUCGAGCACUUGUGCUGCCGUUGG----- --.(((((((((...((....)).)))))))))(((.(((((((-(.(((((.(((..(((.....)))-..))).)))---))..))))))))...)))..........----- ( -39.20, z-score = -2.31, R) >droPer1.super_0 10234080 108 - 11822988 --AACGCAUUAAAUGCGCCCUUGAUUGAUGCGACACUUGCUCGAGGGGUUGCUCAUUAGGUUUAGUGCCGCAGACUGUGCCCCCGAAUGCCACAUGCGUGGUGCUGAGAG----- --..(((((...))))).(((....(((.(((((.(((....)))..))))))))..)))(((((..((((.(..(((((........).))))..)))))..)))))..----- ( -32.70, z-score = -0.03, R) >dp4.chr2 20546893 108 + 30794189 --AACGCAUUAAAUGCGCCCUUGAUUGAUGCGACACUUGCUCGAGGGGUUGCUCAUUAGGUUUAGUGCCACAGACUGUGCCCCCGAAUGCCACAUGCGUGGUGCUGAGAG----- --..(((((...))))).(((....(((.(((((.(((....)))..))))))))..)))(((((..((((.(..(((((........).))))..)))))..)))))..----- ( -33.10, z-score = -0.45, R) >droAna3.scaffold_13340 17083730 105 - 23697760 --AACGCAUAAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUGCUCAUUAGGUAUAGUGCC-CUCUCGGUG---GCAGU---GGCACUUGGGCUUCUGCCGUCGUGC --...((((.....(((((.......)))))(((....(((.((-((((((((.....))))....)))-)))..)))(---((((.---(((......))).)))))))))))) ( -38.20, z-score = -1.23, R) >droEre2.scaffold_4820 6715811 97 + 10470090 --AACGCAUUAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUUCUCAUUAGGUUUAGUGCC-CCGUGCGAG---GCGACUCGAGCACUUGUGCUCC----------- --..((((((((...((....)).))))))))(((..(((((((-(.(((((.(((..(((.....)))-..))).)))---))..))))))))..))).....----------- ( -38.30, z-score = -3.28, R) >droYak2.chr3R 6618185 97 + 28832112 --AACGCAUUAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUUCUCAUUAGGUUUAGUGCC-CCGUGCGAG---GCGACUGGAGCACUUGUGCUCC----------- --..(((......((((((.......))))))...(((((.((.-((((.................)))-))).)))))---)))...((((((....))))))----------- ( -36.33, z-score = -2.51, R) >droSec1.super_22 887892 102 - 1066717 --AACGCAUUAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUUCUCAUUAGGUUUAGUGCC-CG-UGCGAG---GCGACUAGAGCACUUGUGCUACUGUUGC----- --..((((((((...((....)).))))))))...(((((....-((((.................)))-).-.)))))---(((((...((((....))))...)))))----- ( -33.03, z-score = -1.15, R) >droSim1.chr3R 23972340 102 - 27517382 --AACGCAUUAAAUGCGUCCUUGAUUGAUGCGACACUUGCUCGA-GGGCUUCUCAUUAGGUUUAGUGCC-CC-UACGAG---GCGACUCGAACACUUGUGCUGCUGUUGC----- --.((((((...))))))...........((((((...((.(((-((((.................)))-..-..((((---....))))....)))).))...))))))----- ( -28.23, z-score = -0.47, R) >droWil1.scaffold_181089 8955404 104 - 12369635 --AACGCAUUAAAUGCGCUCUUGAUUGAUGCGACACUUGCUUGA-GGGUCUCUCAUUAAGUUUAGUGCCACCAUCAUAU---AAGAAUAAGAGACUGGGUAUGCCCUUCC----- --..(((((...))))).(((((..(((((.(.((((.((((((-(((...))).))))))..)))).)..)))))..)---))))......((..(((....))).)).----- ( -25.10, z-score = -0.76, R) >droVir3.scaffold_12822 3721725 106 - 4096053 AUAAAGCGUUAAAUGCGCCUUUGAUUGAUGCGACACUCGCUCAA-GGGCCACUCAUUAGGUUUAGUGCCACAGUGUCUG-CAACACACACACGAUAACUCCAGCUGCC------- ....(((......((.((((((((.....(((.....)))))))-)))))).((....(((.....)))...((((.((-.....)).))))))........)))...------- ( -25.00, z-score = 0.09, R) >droMoj3.scaffold_6540 17033267 103 - 34148556 AUAAAGCGCUAAAUACGCAUUUGAUUGAUGCGACACUUGCUCAA-GGGCCUCUCAUUAGGUUUAUUGCC-CAGUGUCUG-CCGCAUACACGAUAU--CGCCAGCGAUU------- .....((((.......(((((.....)))))((((((.((....-((((((......))))))...)).-.)))))).)-.))).........((--((....)))).------- ( -25.20, z-score = -0.18, R) >droGri2.scaffold_14906 1321703 106 - 14172833 AUGAAGCGCUAAAUGCGCAUUUGAUUGAUGCGACACUCGCUCAA-GGGCCUCUCAUUAGGUUUAGUGCCACAGUGUCUG-GCACACACACGAUAACACACCGGCACAC------- .....((((.....))))......((((.(((.....)))))))-((((((......)))))).(((((...((((.((-...)).))))(......)...)))))..------- ( -29.20, z-score = -0.82, R) >consensus __AACGCAUUAAAUGCGCCCUUGAUUGAUGCGACACUUGCUCGA_GGGCUUCUCAUUAGGUUUAGUGCC_CAGUGCGAG___GCGAAUACAACACUUGUGCUGCUGUU_______ .....(((((((((..((((....((((.((((...)))))))).))))..((....)))))))))))............................................... (-15.11 = -14.27 + -0.85)

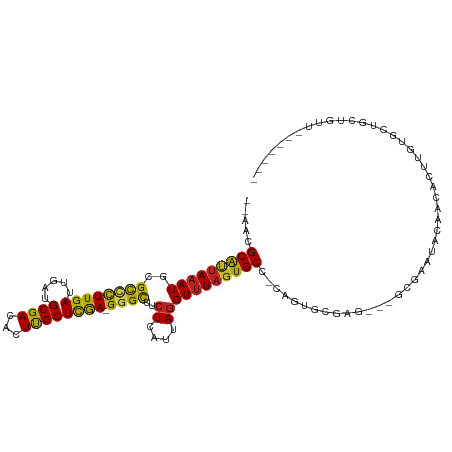
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:14 2011