| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,280,042 – 24,280,151 |
| Length | 109 |
| Max. P | 0.753893 |

| Location | 24,280,042 – 24,280,151 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.43 |
| Shannon entropy | 0.22273 |
| G+C content | 0.46942 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -25.27 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.753893 |
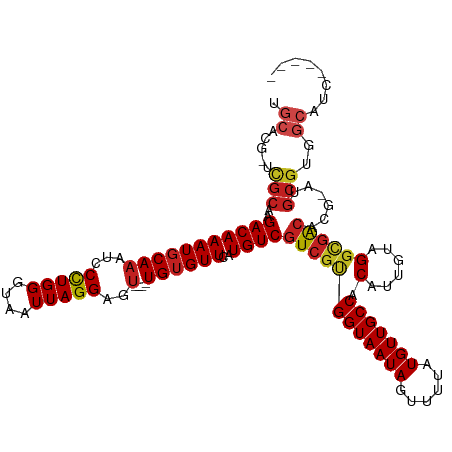
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24280042 109 - 27905053 UGCACG-UCGCAAGACAAAUGCAAAUCCUUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGCGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUC----- (((.((-.((((.(((((((((((.((((.........)))).)----))))))..))))((((((((((((......))))))).(......))))))...-.)))))))))..----- ( -37.10, z-score = -2.29, R) >droSim1.chr3R 23967912 109 - 27517382 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGCGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUC----- (((.((-.((((.(((((((((((...(((((....)))))..)----))))))..))))((((((((((((......))))))).(......))))))...-.)))))))))..----- ( -36.90, z-score = -2.03, R) >droSec1.super_22 883524 109 - 1066717 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGCGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUC----- (((.((-.((((.(((((((((((...(((((....)))))..)----))))))..))))((((((((((((......))))))).(......))))))...-.)))))))))..----- ( -36.90, z-score = -2.03, R) >droYak2.chr3R 6613644 109 + 28832112 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCACGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUC----- ((((.(-((....)))...))))....(((((....))))).((----..(((((.((((((((((((((((......))))))).....)).))))))).)-).)))..))...----- ( -36.60, z-score = -2.19, R) >droEre2.scaffold_4820 6711365 109 + 10470090 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGCGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUC----- (((.((-.((((.(((((((((((...(((((....)))))..)----))))))..))))((((((((((((......))))))).(......))))))...-.)))))))))..----- ( -36.90, z-score = -2.03, R) >droAna3.scaffold_13340 17079802 105 - 23697760 UGCACGUUUGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUG---------- ....((((((((((((((((((((...(((((....)))))..)----))))))..))))...(((((((((......))))))))).))))))))).((((-...))))---------- ( -36.30, z-score = -2.71, R) >dp4.chr2 20542954 115 + 30794189 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACACACGCGCUGCGUGGCAGC (((..(-((((..(((((((((((...(((((....)))))..)----))))))..))))...(((((((((......))))))))).......)))))...(((((...)))))))).. ( -43.30, z-score = -2.47, R) >droPer1.super_0 10230206 115 - 11822988 UGCACG-UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACUCACACGCGCUGCGUGGCAGC (((..(-((((..(((((((((((...(((((....)))))..)----))))))..))))...(((((((((......))))))))).......)))))...(((((...)))))))).. ( -42.80, z-score = -2.50, R) >droWil1.scaffold_181089 8951464 113 - 12369635 UGCACG-CCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGUUGUGUGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG-AUGCGUGGCAUG----- .....(-((((...(((.((((((...(((((....)))))..)))))).)))((.((((((((((((((((......)))))))))......))))))).)-)...)))))...----- ( -39.90, z-score = -1.99, R) >droMoj3.scaffold_6540 17029215 108 - 34148556 -GCAUG-CCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUUGGCGACACG-AUGCGUGGCAAC----- -...((-((((..(((((((((((...(((((....)))))..)----))))))..))))((.(((((((((......)))))))))(((((.(....))))-))))))))))..----- ( -38.20, z-score = -2.33, R) >droVir3.scaffold_12822 3717587 109 - 4096053 UGCAUG-CCGCAAGACAAAUUCAAAUCCCUGGGUAAUUAGGAGU----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUUGGCGACACG-AUGCGUGGCAAC----- ....((-((((............((..(((((....)))))..)----)(((((..((((((((((((((((......)))))))))......))))))).)-))))))))))..----- ( -36.60, z-score = -2.12, R) >droGri2.scaffold_14906 1317711 98 - 14172833 UGCAUG-GCGUAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGA----UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUUGCUUGGUAAC----------------- (((..(-(((...((((...(((.((((((((....))))).))----).)))...))))...(((((((((......))))))))).....))))..)))..----------------- ( -30.50, z-score = -2.45, R) >consensus UGCACG_UCGCAAGACAAAUGCAAAUCCCUGGGUAAUUAGGAGU____UGUGUUCAUGUCGUCGUGGUAAUAGUUUUAUGUUGCCACAUUGUAGGCGACACG_AUGCGUGGCAUC_____ .((.....(((..((((((((((...((((((....))))).).....))))))..))))((((((((((((......))))))).(......))))))......)))..))........ (-25.27 = -25.77 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:13 2011