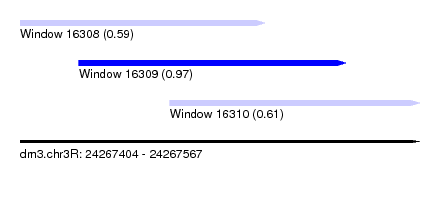
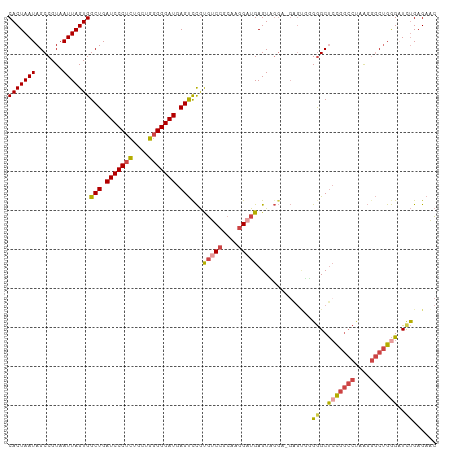
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,267,404 – 24,267,567 |
| Length | 163 |
| Max. P | 0.968865 |

| Location | 24,267,404 – 24,267,504 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.32 |
| Shannon entropy | 0.60101 |
| G+C content | 0.57574 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
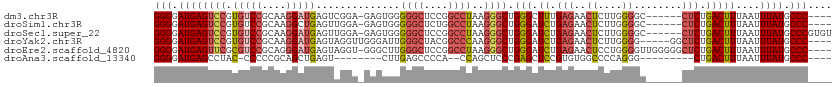
>dm3.chr3R 24267404 100 + 27905053 ----GCGCU-CCUGUUGUUCCUGGGC-CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUCGGAGAGUG- ----((.((-((.((((((..(((..-..)))..))))))........(.((((.(((((((....))))))).)))))..((((....)))).....)))).)).- ( -39.90, z-score = -2.10, R) >droSim1.chr3R 23955117 100 + 27517382 ----GCGCU-CCUGUUGUUCCUGGGC-CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGCUGAGUUGGAGAGUG- ----((.((-((.((((((..(((..-..)))..)))))).....(((((((((.(((((((....))))))).))))......(....))))))...)))).)).- ( -37.00, z-score = -1.15, R) >droSec1.super_22 870910 100 + 1066717 ----GCGCU-CCUGUUAUUCCGGGGC-CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUUGGAGAGUG- ----((.((-((....((((((((..-..((((((((.......))))))))((.(((((((....))))))).)))))).((((....)))))))).)))).)).- ( -41.40, z-score = -2.39, R) >droYak2.chr3R 6600740 101 - 28832112 ----GCGCU-CCUGUUGUUCUUCGGC-CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCGCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGUUGGGAU ----.(.(.-(((((..((((((((.-..((((((((.......))))))))((.(((((.(....).))))).)).......))).)))))..).))))..).).. ( -34.10, z-score = -0.02, R) >droEre2.scaffold_4820 6697614 100 - 10470090 ----GCGCU-CCUGUUGUUCUUGUGC-CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCAGCUGGGGGAUGAGUUCGCGUCCGCAGGGAUGAGUAGGUGGGCU- ----((.(.-(((((..((((((((.-.(((((((((.......))))))))((.(((((((....))))))).)).....)..))))))))..).)))).).)).- ( -42.60, z-score = -2.20, R) >droAna3.scaffold_13340 17067254 92 + 23697760 ----GCGCU-CCUGUUGUUGCU-GCCGUUCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUUGAC-GGGGGAUGAGCCUAC-CCCCCGCAGCUGAGUCUU------- ----..(((-(........(((-((......((((((.......))))))((((.((((((....-.)))))).))))...-.....))))).))))...------- ( -30.00, z-score = -1.35, R) >dp4.chr2 20528908 97 - 30794189 CCUCUCGCU-CCUUCAGCUGUUUGCCGCUCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUUGGC-GGGGGACCACUUCGC-UCCCCAGCUCCCAGUUGC------- .........-....((((((...((((((((((((((.......))))))))..........)))-)(((((.(.....).-))))).))...)))))).------- ( -28.90, z-score = -0.74, R) >droPer1.super_0 10216267 97 + 11822988 CCUCUCGCU-CCUUCAGUUGUUUGCCGCUCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUUGGC-GGGGGACCACUUCGC-UCCCCAGCUCCCAGUUGC------- .((((((((-...(((((..(((((.(((........))).))))...)..)))))......)))-)))))..........-....((((.....)))).------- ( -25.20, z-score = 0.25, R) >droWil1.scaffold_181089 8937832 87 + 12369635 ---CCUGCUACUUGCUACUUCUGCCUCUUUCACUAAUAGCCGUAAUUAGUGGCCGAUCC---AGUUGAGAAUGUAGGGCGUAGGCGGAGGGUA-------------- ---...((.....))..((((((((((...(((((((.......)))))))(((..(.(---(........)).).)))).)))))))))...-------------- ( -24.70, z-score = 0.28, R) >consensus ____GCGCU_CCUGUUGUUCCUGGGC_CCCCACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUUGG___G___ ....((((.....................((((((((.......))))))))....((((((....)))))).......))))........................ (-15.38 = -15.97 + 0.58)

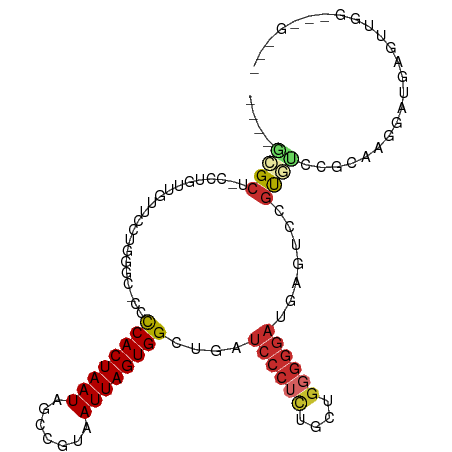
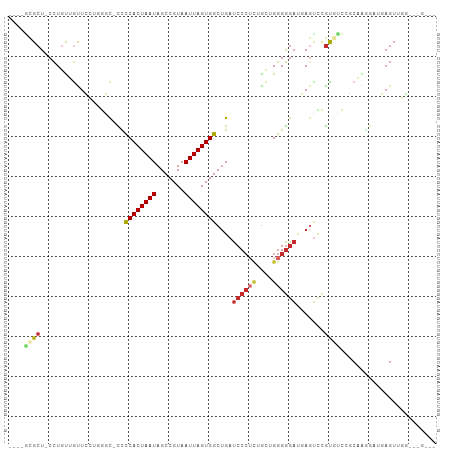
| Location | 24,267,428 – 24,267,537 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Shannon entropy | 0.35021 |
| G+C content | 0.58587 |
| Mean single sequence MFE | -43.80 |
| Consensus MFE | -32.27 |
| Energy contribution | -33.25 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.968865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
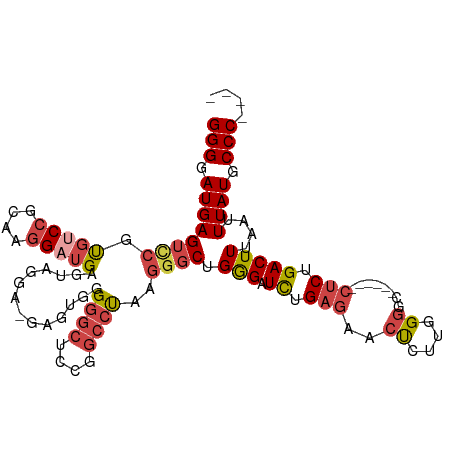
>dm3.chr3R 24267428 109 + 27905053 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUCGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGCUUUGAGAAC ((((.....(((......(.((((.(((((((....))))))).)))))..((((....))))....))).-.))))(((((.((((((....)))))))))))...... ( -48.70, z-score = -2.83, R) >droSim1.chr3R 23955141 109 + 27517382 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGCUGAGUUGGA-GAGUGGGGGCUCUGGCCUAAGGGCUGGGAUCUGAGAAC ((((.....(((...(((((((((.(((((((....))))))).))))......(....))))))..))).-.)))).(((..(..(((....)))..)..)))...... ( -42.00, z-score = -1.06, R) >droSec1.super_22 870934 109 + 1066717 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUUGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAAC ((((..(((((.......(.((((.(((((((....))))))).)))))..((((....))))).))))..-.)))).(((..((((((....))))))..)))...... ( -46.80, z-score = -2.66, R) >droYak2.chr3R 6600764 110 - 28832112 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCGCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGUUGGGAUUGGGCUACGGCCCAAGGGCUGGGAUCUGAGAAC (((((((.......)))))))...((((((.(((((...(((....)))..((((....)))).)))))....((.((((((....))))))...))))))))....... ( -44.30, z-score = -2.21, R) >droEre2.scaffold_4820 6697638 109 - 10470090 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCAGCUGGGGGAUGAGUUCGCGUCCGCAGGGAUGAGUAGGU-GGGCUUGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAAC (((((((.......)))))))(((.(((((((....))))))).)))...(((((....))))).......-(..((..((..((((((....))))))..))..))..) ( -45.90, z-score = -1.94, R) >droAna3.scaffold_13340 17067278 98 + 23697760 CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUUGAC-GGGGGAUGAGCCUACCCCC-CGCAGCUGAGU--------CUUGAGCCCCA--CCAGCUCCCGAGCUCCGUGUGG .((((((.......))))))((((.((((((....-.)))))).))))......(-((((.(.(((.--------(((((((....--...))))..)))))).)))))) ( -35.10, z-score = -1.99, R) >consensus CACUAAUAGCCGUAAUUAGUGGCUGAUCCCUCUGCUGGGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGA_GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAAC (((((((.......)))))))(((.(((((((....))))))).)))...(((((....)))))..............((..(((((((....)))))))..))...... (-32.27 = -33.25 + 0.98)

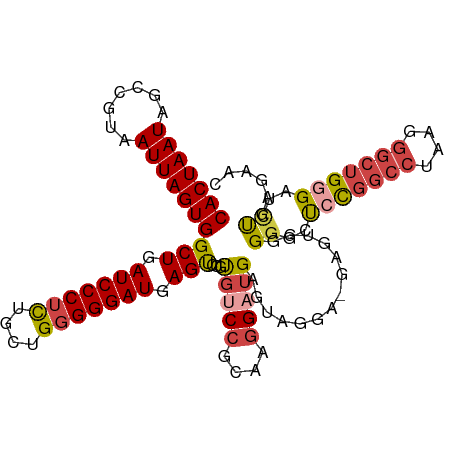
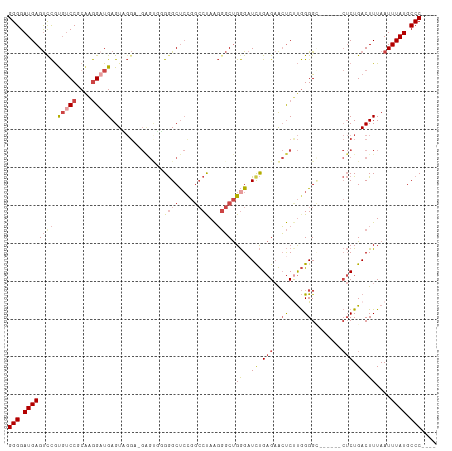
| Location | 24,267,465 – 24,267,567 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.76 |
| Shannon entropy | 0.41890 |
| G+C content | 0.59196 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -21.82 |
| Energy contribution | -23.85 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.613886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24267465 102 + 27905053 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUCGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGCUUUGAGAACUCUUGGGGC------CUCUGACUUUAAUUUAUGCCC---- (((.((((((....((((....))))((((((((-(..........((((((....))))))((((..((....))..))))------)))))))))..)))))).)))---- ( -47.40, z-score = -2.89, R) >droSim1.chr3R 23955178 102 + 27517382 GGGGAUGAGUCCGUGUCCGCAAGGCUGAGUUGGA-GAGUGGGGGCUCUGGCCUAAGGGCUGGGAUCUGAGAACUCUUGGGGC------CUCUGACUUUAAUUUAUGCCC---- (((.(((....))).)))....((((((((((((-(..(((((((((..(((....)))..)).((..((....))..))))------))))).)))))))))).))).---- ( -41.20, z-score = -1.36, R) >droSec1.super_22 870971 106 + 1066717 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUUGGA-GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCUUGGGGC------CUCUGACUUUAAUUUAUGCCCGUGU (((.(((((((((.((((....))))....)))(-(((((((((((((((((....))))))).((..((....))..))))------)))).))))).)))))).))).... ( -47.40, z-score = -2.80, R) >droYak2.chr3R 6600801 104 - 28832112 GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGUUGGGAUUGGGCUACGGCCCAAGGGCUGGGAUCUGAGAACUCUUGGGG-----GGCUCUGACUUUAAUUUAUGCCC---- (((.((((((....((((....))))....((((..(..((((((....)))))).((((....((..((....))..)).-----)))))..))))..)))))).)))---- ( -40.30, z-score = -1.52, R) >droEre2.scaffold_4820 6697675 108 - 10470090 GGGGAUGAGUUCGCGUCCGCAGGGAUGAGUAGGU-GGGCUUGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCCUGGGGUUGGGGGCUCUGACUUUAAUUUAUGCCC---- (((.(((((((..(((((....)))))..((((.-(..((..((..((((((....))))))..))..))..).))))((((..(.....)..)))).))))))).)))---- ( -45.20, z-score = -1.40, R) >droAna3.scaffold_13340 17067314 89 + 23697760 GGGGAUGAGCCUAC-CCCCCGCAGCUGAGU--------CUUGAGCCCCA--CCAGCUCCCGAGCUCCGUGUGGCCCCAGGG---------CUGACUUUAAUUUAUGCCC---- ((((..........-.))))(((..(.(((--------((((.(((.((--(.((((....))))..))).)))..)))))---------)).)..........)))..---- ( -27.10, z-score = -0.03, R) >consensus GGGGAUGAGUCCGUGUCCGCAAGGAUGAGUAGGA_GAGUGGGGGCUCCGGCCUAAGGGCUGGGAUCUGAGAACUCUUGGGGC______CUCUGACUUUAAUUUAUGCCC____ (((.((((((....((((....))))((((.....((((..((..(((((((....)))))))..))....)))).(((((.......)))))))))..)))))).))).... (-21.82 = -23.85 + 2.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:10 2011