| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,267,051 – 24,267,146 |
| Length | 95 |
| Max. P | 0.858958 |

| Location | 24,267,051 – 24,267,146 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.45 |
| Shannon entropy | 0.56439 |
| G+C content | 0.53676 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -15.16 |
| Energy contribution | -17.11 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.858958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24267051 95 - 27905053 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGUCGAGGCCG--AAGGGCCAUCU-CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG--- -------.......(((((((................((((((.((((.--...)))).((.-.....)).))))))..((((((......))))))))))))).--- ( -36.20, z-score = -2.26, R) >droSim1.chr3R 23954773 95 - 27517382 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGUCGAGGCCG--AAGGGCCAUCU-CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG--- -------.......(((((((................((((((.((((.--...)))).((.-.....)).))))))..((((((......))))))))))))).--- ( -36.20, z-score = -2.26, R) >droSec1.super_22 870565 95 - 1066717 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGUCGAGGCCG--AAGGGCCAUCU-CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG--- -------.......(((((((................((((((.((((.--...)))).((.-.....)).))))))..((((((......))))))))))))).--- ( -36.20, z-score = -2.26, R) >droYak2.chr3R 6600397 95 + 28832112 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGCCGAGGCCG--AAGGGCCAUCU-CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG--- -------.......(((((((................((((((.((((.--...)))).((.-.....)).))))))..((((((......))))))))))))).--- ( -38.90, z-score = -2.84, R) >droEre2.scaffold_4820 6697274 95 + 10470090 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGUCGAGGCCG--AAGGGCCAUCG-CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG--- -------.......(((((((...(((((....)))))(..((.((((.--...)))).(((-(((((((.......)))))))))).....))..)))))))).--- ( -39.00, z-score = -2.76, R) >droAna3.scaffold_13340 17066903 93 - 23697760 -------CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGCCCGGCCGAGUAAGGGCCGUUU-CAGCUGGC--GUCA--GACUGCGCCGCAGCGGUUGCCCUUUG--- -------.....(((((.(((................(((..(((((.......)))))..)-))((((((--((..--....)))))..)))....))))))))--- ( -30.80, z-score = -0.16, R) >dp4.chr2 20528534 96 + 30794189 -------CCGGCCGAAGUGGCUUAAUAAAAUAAUUUAUGAUUUCGGCCC-UGAGGCACAUUU----UUGGGGGGCACCCCACUGCGACGCAGCGGUUGCCACUUGUCC -------..(((((((((.(...(((......)))..).))))))))).-...((((.....----..((((....))))(((((......)))))))))........ ( -33.80, z-score = -1.31, R) >droPer1.super_0 10215886 96 - 11822988 -------CCGGCCGAAGUGGCUUAAUAAAAUAAUUUAUGAUUUCGGCCC-UGAGGCACAUUU----UUGGGGGGCACCCCACUGCGACGCAGCGGUUGCCACUUGUCC -------..(((((((((.(...(((......)))..).))))))))).-...((((.....----..((((....))))(((((......)))))))))........ ( -33.80, z-score = -1.31, R) >droWil1.scaffold_181089 8937396 101 - 12369635 GGGGCCCCCGGUCGAAGUGGCUUAAUAAAAUAAUUUAUGUUGUCGGCCAUUGAGUUGAGAAU-GGCGCGUCCGGUUG--AGUUGUGGCUCUUUGCUUUUUGUUU---- (((((((((((.((...((((...(((((....)))))...))))((((((........)))-))).)).))))...--....).)))))).............---- ( -27.21, z-score = -0.08, R) >droVir3.scaffold_12822 3703715 84 - 4096053 -------CCAGCCAAAGUGGCUUAAUAAAAUAAUUUAUGGUUUGGCAA---AAUGCCCAUUUACCGCCACACAGCCCCAACCUUUGUCUUACUC-------------- -------.........(((((...(((((....)))))(((..(((..---...))).....))))))))........................-------------- ( -15.60, z-score = -0.69, R) >consensus _______CCGUCCGAAGUGGCUUAAUAAAAUAAUUUAUGGUCGAGGCCG__AAGGGCCAUCU_CAGUUGGCCGGCCACCAACUGCGACGCAGCGGUUGCCACUUG___ ..............(((((((.......................(((((......((((........)))))))))....(((((......))))).))))))).... (-15.16 = -17.11 + 1.95)


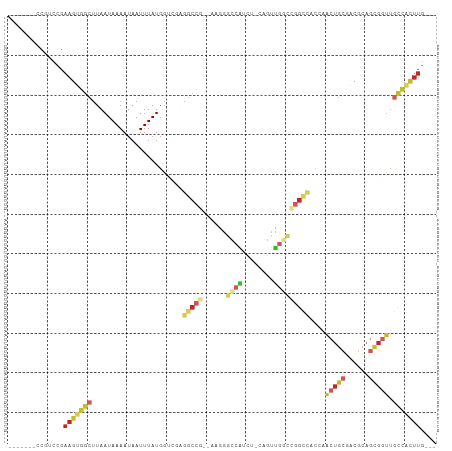
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:08 2011