| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,266,944 – 24,267,035 |
| Length | 91 |
| Max. P | 0.874303 |

| Location | 24,266,944 – 24,267,035 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 66.83 |
| Shannon entropy | 0.68313 |
| G+C content | 0.44313 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.866050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24266944 91 + 27905053 -CGAAAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG-GGCAGCAACCAUCCAGAACUCCC-------------------- -.((((...(((((((((........)))))))))((((.....))))....))))..........(((-((.........))))).......-------------------- ( -20.40, z-score = -0.64, R) >droSim1.chr3R 23954668 89 + 27517382 ---CGAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG-GGCAGCAACCACCCAGAACUCCC-------------------- ---.((((((((((((((........)))))))))((((.....))))..................(((-((.........))))))))))..-------------------- ( -25.40, z-score = -2.42, R) >droSec1.super_22 870458 91 + 1066717 -CGAAAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG-GGCAGCAACCACCCAGAACUCCC-------------------- -.((((...(((((((((........)))))))))((((.....))))....))))..........(((-((.........))))).......-------------------- ( -23.20, z-score = -1.57, R) >droYak2.chr3R 6600283 98 - 28832112 -CUAAAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG-GGCAGCAACCACCCAGAGAAAUUGGGGCCC------------- -........(((((((((........)))))))))((((.....))))...((..((....((...(((-((.........)))))))....))..))..------------- ( -25.60, z-score = -0.66, R) >droEre2.scaffold_4820 6697164 94 - 10470090 -CUAAAGUUGAGUCAAAGGUUUUUCGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG-GGCAGCAACCACCCAGAACUGCCCCC----------------- -....(((((((((((((........)))))))))((((.....))))................))))(-(((((.............))))))..----------------- ( -25.22, z-score = -2.19, R) >droAna3.scaffold_13340 17066793 85 + 23697760 -CAAAAGUUGAGUCAAAGGUUUU-CGCUUUGACUCGUAAUUAAUUUGCUUUGUUCCGUCCGCUUGGCCG-GGCAGCCAUCAGCGGGUG------------------------- -((((....(((((((((.....-..)))))))))((((.....))))))))...(..(((((((((..-....))))..)))))..)------------------------- ( -28.50, z-score = -2.45, R) >dp4.chr2 20528376 103 - 30794189 -UAAAAGUUGAGUCAAAGGUUUU-CUCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUGCGCUUCGCUG-GGCAAUAGACAUAGGCCAUAGACGGUCCCCCGCGUC------- -........((((((((((....-.))))))))))...........(((((((((..(((.(......)-.)))..))))).))))....((((........))))------- ( -26.90, z-score = -1.17, R) >droPer1.super_0 10215730 101 + 11822988 -CAAAAGUUGAGUCAAAGGUUUU-CUCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUGCGCUUCGCUG-GGCAAUAGACAUAGGCAAUAGACGGUCCCCCGUC--------- -........((((((((((....-.)))))))))).........(((((((((((..(((.(......)-.)))..))))).))))))..(((((....)))))--------- ( -32.30, z-score = -3.49, R) >droWil1.scaffold_181089 8937255 92 + 12369635 UGUUUGGCCCAGUCAAAAGGUU--GCCUUUGACUUGCAAUUAAUUCGCUUUGUUUUUCACGCUUGACUG-GACAACACAAACACAGAUAGAGCGA------------------ ......((..(((((((.((..--.))))))))).)).......(((((((((...(((....)))(((-.............))))))))))))------------------ ( -19.02, z-score = 0.21, R) >droMoj3.scaffold_6540 17014170 109 + 34148556 -UGUUCAUGGAGUCAAA-GGUU--CGCUUUGACUUGCAAUUAAUUUGCUUUGUUUUUUGCACUUGACUGCGCUUGGACAGACUUACAAGCUGCCAGGCAGAUAGAUAGAAGGA -(.(((...((((((((-(...--..)))))))))((((.....))))(((((((...(((......)))((((((.(((.((....))))))))))))))))))..))).). ( -31.80, z-score = -1.38, R) >droVir3.scaffold_12822 3703560 105 + 4096053 -UGUUCAUGGAGUCAAA-GGUU--CGCUUUGACUUGCAAUUAAUUUGCUUUGUUUUUUGCACUUGACUGCGCCCGGACAGACUUACAAGCGCUCAUCCGGACAGAUAGA---- -(((((...((((((((-(...--..)))))))))((((..(((.......)))..))))...(((..((((..(..........)..)))))))...)))))......---- ( -24.40, z-score = 0.15, R) >droGri2.scaffold_14906 1304625 105 + 14172833 -UUUUAGUAAAGUCAAA-GGUU--CCCUUUGACUUGCAAUUAAUUUGCUUUGUUUUUUGCACUUGACUGCGCCUGGACAAACAGACAAGACGUCGAACAGACAGAAAGA---- -.....((((.((((((-((..--.))))))))))))......((((.(((((((..((((......))))...)))))))))))......(((.....))).......---- ( -25.30, z-score = -1.23, R) >consensus _CGAAAGUUGAGUCAAAGGUUUU_CGCUUUGACUCGUAAUUAAUUUGCUUUGUUUCAUCCGCUUGACUG_GGCAGCAAACACCCAGAACACCC____________________ .........((((((((.((.....))))))))))((((.....))))................................................................. (-10.38 = -10.26 + -0.12)



| Location | 24,266,944 – 24,267,035 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 66.83 |
| Shannon entropy | 0.68313 |
| G+C content | 0.44313 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -8.03 |
| Energy contribution | -7.98 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24266944 91 - 27905053 --------------------GGGAGUUCUGGAUGGUUGCUGCC-CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUUUCG- --------------------(..((((...(((..(((((...-...(((......))).....)))))....)))..(((((((((........)))))))))))))..).- ( -24.30, z-score = -1.61, R) >droSim1.chr3R 23954668 89 - 27517382 --------------------GGGAGUUCUGGGUGGUUGCUGCC-CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUCG--- --------------------..((((((((((..(...)..))-)))....((............))...........(((((((((........)))))))))))))).--- ( -27.40, z-score = -2.43, R) >droSec1.super_22 870458 91 - 1066717 --------------------GGGAGUUCUGGGUGGUUGCUGCC-CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUUUCG- --------------------(..(((((((((..(...)..))-)))....((............))...........(((((((((........)))))))))))))..).- ( -28.50, z-score = -2.59, R) >droYak2.chr3R 6600283 98 + 28832112 -------------GGGCCCCAAUUUCUCUGGGUGGUUGCUGCC-CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUUUAG- -------------((((..((((..(.....)..))))..)))-)......((............))...........(((((((((........)))))))))........- ( -24.00, z-score = -0.69, R) >droEre2.scaffold_4820 6697164 94 + 10470090 -----------------GGGGGCAGUUCUGGGUGGUUGCUGCC-CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCGAAAAACCUUUGACUCAACUUUAG- -----------------..(((((((.((....))..))))))-)......((............))...........(((((((((........)))))))))........- ( -26.60, z-score = -1.38, R) >droAna3.scaffold_13340 17066793 85 - 23697760 -------------------------CACCCGCUGAUGGCUGCC-CGGCCAAGCGGACGGAACAAAGCAAAUUAAUUACGAGUCAAAGCG-AAAACCUUUGACUCAACUUUUG- -------------------------...(((((..(((((...-.)))))))))).......................(((((((((.(-....))))))))))........- ( -27.40, z-score = -3.69, R) >dp4.chr2 20528376 103 + 30794189 -------GACGCGGGGGACCGUCUAUGGCCUAUGUCUAUUGCC-CAGCGAAGCGCAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGAG-AAAACCUUUGACUCAACUUUUA- -------...((((....))))((.((...(((((((.((((.-..)))))).)))))...)).))............(((((((((..-.....)))))))))........- ( -26.00, z-score = -1.43, R) >droPer1.super_0 10215730 101 - 11822988 ---------GACGGGGGACCGUCUAUUGCCUAUGUCUAUUGCC-CAGCGAAGCGCAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGAG-AAAACCUUUGACUCAACUUUUG- ---------(((((....)))))..((((...(((....(((.-(......).)))....)))..)))).........(((((((((..-.....)))))))))........- ( -30.40, z-score = -3.66, R) >droWil1.scaffold_181089 8937255 92 - 12369635 ------------------UCGCUCUAUCUGUGUUUGUGUUGUC-CAGUCAAGCGUGAAAAACAAAGCGAAUUAAUUGCAAGUCAAAGGC--AACCUUUUGACUGGGCCAAACA ------------------............((((((....(((-((((((((.............((((.....))))..((.....))--.....))))))))))))))))) ( -22.80, z-score = -0.94, R) >droMoj3.scaffold_6540 17014170 109 - 34148556 UCCUUCUAUCUAUCUGCCUGGCAGCUUGUAAGUCUGUCCAAGCGCAGUCAAGUGCAAAAAACAAAGCAAAUUAAUUGCAAGUCAAAGCG--AACC-UUUGACUCCAUGAACA- ...............((.((((((((....)).))).))).))(((((....(((..........))).....))))).((((((((..--...)-))))))).........- ( -24.10, z-score = -1.28, R) >droVir3.scaffold_12822 3703560 105 - 4096053 ----UCUAUCUGUCCGGAUGAGCGCUUGUAAGUCUGUCCGGGCGCAGUCAAGUGCAAAAAACAAAGCAAAUUAAUUGCAAGUCAAAGCG--AACC-UUUGACUCCAUGAACA- ----.......((((((((..((........))..))))))))(((((....(((..........))).....))))).((((((((..--...)-))))))).........- ( -31.30, z-score = -2.59, R) >droGri2.scaffold_14906 1304625 105 - 14172833 ----UCUUUCUGUCUGUUCGACGUCUUGUCUGUUUGUCCAGGCGCAGUCAAGUGCAAAAAACAAAGCAAAUUAAUUGCAAGUCAAAGGG--AACC-UUUGACUUUACUAAAA- ----.......(((.....)))((((((.((((..((....)))))).)))).))..........((((.....))))((((((((((.--..))-))))))))........- ( -26.20, z-score = -1.95, R) >consensus ____________________GGCAGUUCUGGGUGUUUGCUGCC_CAGUCAAGCGGAUGAAACAAAGCAAAUUAAUUACGAGUCAAAGCG_AAAACCUUUGACUCAACUUUAG_ ...................................................((............))...........((((((((..........))))))))......... ( -8.03 = -7.98 + -0.05)

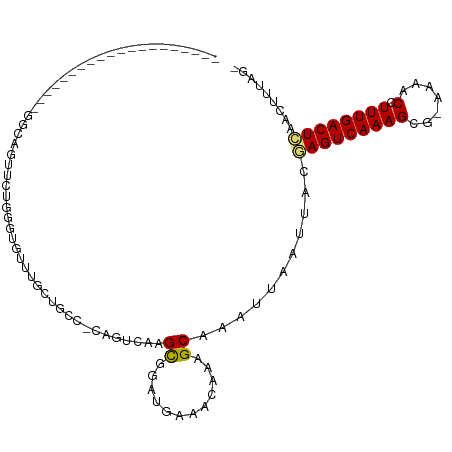

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:07 2011