| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,879,431 – 8,879,523 |
| Length | 92 |
| Max. P | 0.991417 |

| Location | 8,879,431 – 8,879,523 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 66.38 |
| Shannon entropy | 0.60100 |
| G+C content | 0.33918 |
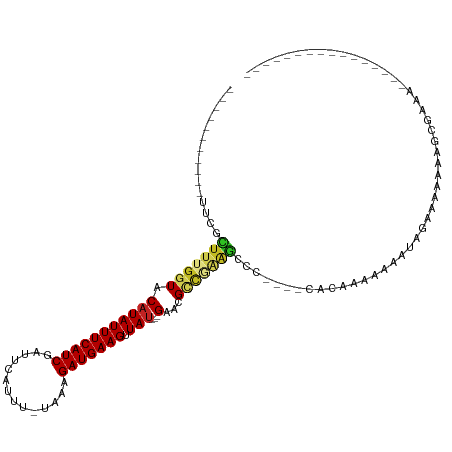
| Mean single sequence MFE | -18.69 |
| Consensus MFE | -8.65 |
| Energy contribution | -8.61 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
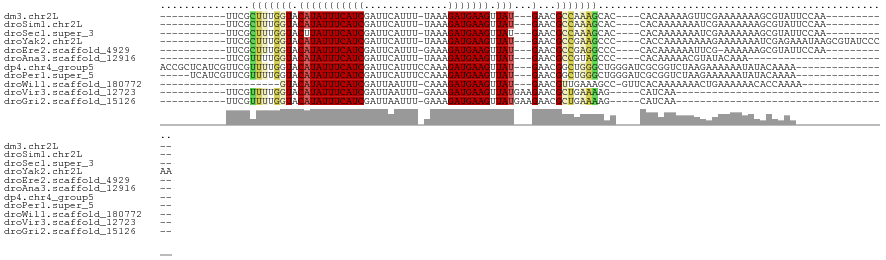
>dm3.chr2L 8879431 92 - 23011544 -----------UUCGCUUUGGUACAUAUUUCAUCGAUUCAUUU-UAAAGAUGAAGUUAU---GAACGCCAAAGCAC----CACAAAAAGUUCGAAAAAAAGCGUAUUCCAA----------- -----------...((((((((.(((((((((((.........-....))))))).)))---)...))))))))..----...............................----------- ( -20.82, z-score = -3.31, R) >droSim1.chr2L 8658257 92 - 22036055 -----------UUCGCUUUGGUACAUAUUUCAUCGAUUCAUUU-UAAAGAUGAAGUUAU---GAACGCCAAAGCAC----CACAAAAAAAUCGAAAAAAAGCGUAUUCCAA----------- -----------...((((((((.(((((((((((.........-....))))))).)))---)...))))))))..----...............................----------- ( -20.82, z-score = -3.64, R) >droSec1.super_3 4353464 92 - 7220098 -----------UUCGCUUUGGUACUUAUUUCAUCGAUUCAUUU-UAAAGAUGAAGUUAU---GAACGCCAAAGCAC----CACAAAAAAAUCGAAAAAAAGCGUAUUCCAA----------- -----------...((((((((....((((((((.........-....))))))))...---....))))))))..----...............................----------- ( -18.72, z-score = -2.67, R) >droYak2.chr2L 11527224 103 - 22324452 -----------UUCGCUUUGGUACAUAUUUCAUCGAUUCAUUU-UAAAGAUGAAGUUAU---GAACGCCGAAGCCC----CACCAAAAAAAAGAAAAAAAUCGAGAAAUAAGCGUAUCCCAA -----------(((((((((((.(((((((((((.........-....))))))).)))---)...))))))))..----............((......))..)))............... ( -19.72, z-score = -2.68, R) >droEre2.scaffold_4929 9472809 91 - 26641161 -----------UUCGCUUUGGUACAUAUUUCAUCGAUUCAUUU-GAAAGAUGAAGUUAU---GAACGCCGAGGCCC----CACAAAAAAUUCG-AAAAAAGCGUAUUCCAA----------- -----------...((((((((.(((((((((((..(((....-))).))))))).)))---)...))))))))..----.............-.................----------- ( -21.60, z-score = -2.41, R) >droAna3.scaffold_12916 10544064 79 - 16180835 -----------UUCGUUUUGGUACAUAUUUCAUCGAUUCAUUU-UAAAGAUGAAGUUAU---GAACGCCGUAGCCC----CACAAAAACGUAUACAAA------------------------ -----------..(((((((((.(((((((((((.........-....))))))).)))---)...)))((.....----.)).))))))........------------------------ ( -14.12, z-score = -1.66, R) >dp4.chr4_group5 748356 103 - 2436548 ACCGCUCAUCGUUCGUUUUGGUACAUAUUUCAUCGAUUCAUUUCCAAAGAUGAAGUUAU---GAACGGCUGGGCUGGGAUCGCGGUCUAAGAAAAAAUAUACAAAA---------------- .(((((((((((((((..........((((((((..............)))))))).))---)))))).))))).))(((....)))...................---------------- ( -24.34, z-score = -1.05, R) >droPer1.super_5 5154484 98 - 6813705 -----UCAUCGUUCGUUUUGGUACAUAUUUCAUCGAUUCAUUUCCAAAGAUGAAGUUAU---GAACGGCUGGGCUGGGAUCGCGGUCUAAGAAAAAAUAUACAAAA---------------- -----...((((((((..........((((((((..............)))))))).))---)))))).(((((((......))))))).................---------------- ( -20.14, z-score = -0.53, R) >droWil1.scaffold_180772 8698058 82 + 8906247 --------------------GUACAUAUUUCAUCGAUUAAUUU-CAAAGAUGAAGUUAU---GAACGUUGAAAGCC-GUUCACAAAAAAACUGAAAAAACACCAAAA--------------- --------------------.......(((((.....((((((-((....))))))))(---(((((.(....).)-))))).........)))))...........--------------- ( -13.50, z-score = -2.06, R) >droVir3.scaffold_12723 4900485 69 - 5802038 -----------UUCGUUUUGGUACAUAUUUCAUCGAUUAAUUU-GAAAGAUGAAGUUAUGAAGAACGCUGAAAAG-----CAUCAA------------------------------------ -----------...(((((....(((((((((((.........-....))))))).)))).)))))(((....))-----).....------------------------------------ ( -15.92, z-score = -2.25, R) >droGri2.scaffold_15126 7901592 69 - 8399593 -----------UUCGUUUUGGUACAUAUUUCAUCGAUUAAUUU-GAAAGAUGAAGUUAUGAAGAACGCUGAAAAG-----CAUCAA------------------------------------ -----------...(((((....(((((((((((.........-....))))))).)))).)))))(((....))-----).....------------------------------------ ( -15.92, z-score = -2.25, R) >consensus ___________UUCGCUUUGGUACAUAUUUCAUCGAUUCAUUU_UAAAGAUGAAGUUAU___GAACGCCGAAGCCC____CACAAAAAAAUAGAAAAAAAGCGAAA________________ ...............(((((((......((((((..............))))))(((......))))))))))................................................. ( -8.65 = -8.61 + -0.04)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:14 2011