| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,250,841 – 24,250,934 |
| Length | 93 |
| Max. P | 0.951917 |

| Location | 24,250,841 – 24,250,934 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
| Shannon entropy | 0.37482 |
| G+C content | 0.59217 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -23.44 |
| Energy contribution | -24.83 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
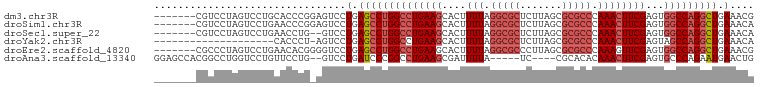
>dm3.chr3R 24250841 93 + 27905053 -------CGUCCUAGUCCUGCACCCGGAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACG -------.....(((..((.(....).))..)))((((((((((((((....(((.(((((.......))))).))))))))...)))))))))...... ( -34.80, z-score = -1.41, R) >droSim1.chr3R 23938564 93 + 27517382 -------CGUCCUAGUCCUGAACCCGGAGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACA -------..(((..((.....))..)))((..(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).).)). ( -33.80, z-score = -1.34, R) >droSec1.super_22 854505 91 + 1066717 -------CGUCCUAGUCCUGAACCUG--GUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACA -------.(..((((........)))--)..)(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).).... ( -34.60, z-score = -1.88, R) >droYak2.chr3R 6583711 79 - 28832112 --------------------CACCCU-AGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUAGCCAGGCUGAAACA --------------------......-.((..(.((((((((((((((....(((.(((((.......))))).))))))).))..)))))))).).)). ( -28.60, z-score = -2.05, R) >droEre2.scaffold_4820 6679212 93 - 10470090 -------CGCCCUAGUCCUGAACACGGGGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCCCUUAGCGCGCCCAAAGUUCGAGUGGCCAGGCUGAAACG -------.(((((.((.......)))))))..(.(((((((((....(.(((((..(((((.......))))).))))).)....))))))))).).... ( -37.60, z-score = -1.56, R) >droAna3.scaffold_13340 17050569 89 + 23697760 GGAGCCACGGCCUGGUCCUGUUCCUG--GUCCUGAUCCCGGCCUGAAGCGAUUUUA-----UC----CGCACACAAACUUCGAGUGCCCAGAAUGAACUG ((.((.((((((.((.((.......)--).)).(....)))))....(((......-----..----))).............))))))........... ( -19.90, z-score = 1.05, R) >consensus _______CGUCCUAGUCCUGAACCCG_AGUCCUGAGCCUGGCCUGAAGCACUUUUAGGCGCUCUUAGCGCGCCCAAACUUCGAGUGGCCAGGCUGAAACA ................................(.((((((((((((((....(((.(((((.......))))).))))))))...))))))))).).... (-23.44 = -24.83 + 1.39)

| Location | 24,250,841 – 24,250,934 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.73 |
| Shannon entropy | 0.37482 |
| G+C content | 0.59217 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -29.42 |
| Energy contribution | -30.23 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
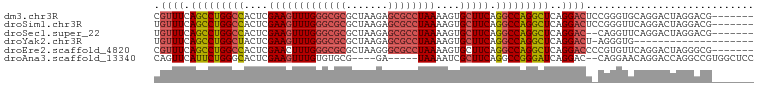
>dm3.chr3R 24250841 93 - 27905053 CGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUCCGGGUGCAGGACUAGGACG------- .((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..))))(((.(((.....))).)))..------- ( -41.70, z-score = -2.26, R) >droSim1.chr3R 23938564 93 - 27517382 UGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACUCCGGGUUCAGGACUAGGACG------- .((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..))))(((.((((...)))).)))..------- ( -41.40, z-score = -2.56, R) >droSec1.super_22 854505 91 - 1066717 UGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGAC--CAGGUUCAGGACUAGGACG------- .((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..))))--...((((.......)))).------- ( -39.20, z-score = -2.46, R) >droYak2.chr3R 6583711 79 + 28832112 UGUUUCAGCCUGGCUACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACU-AGGGUG-------------------- .((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..)))).-......-------------------- ( -35.40, z-score = -2.53, R) >droEre2.scaffold_4820 6679212 93 + 10470090 CGUUUCAGCCUGGCCACUCGAACUUUGGGCGCGCUAAGGGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACCCCGUGUUCAGGACUAGGGCG------- .((((.(((((((((......(((((((((((.(...).)))))).)))))......)))))))))..))))(((..(((...)))..)))..------- ( -39.30, z-score = -1.29, R) >droAna3.scaffold_13340 17050569 89 - 23697760 CAGUUCAUUCUGGGCACUCGAAGUUUGUGUGCG----GA-----UAAAAUCGCUUCAGGCCGGGAUCAGGAC--CAGGAACAGGACCAGGCCGUGGCUCC ........((((.((((.........)))).))----))-----.......(((...((((.((.((..(..--......)..)))).))))..)))... ( -25.30, z-score = 0.75, R) >consensus CGUUUCAGCCUGGCCACUCGAAGUUUGGGCGCGCUAAGAGCGCCUAAAAGUGCUUCAGGCCAGGCUCAGGACU_CGGGUUCAGGACUAGGACG_______ .((((.(((((((((....(((((((((((((.......))))))))....))))).)))))))))..))))............................ (-29.42 = -30.23 + 0.81)
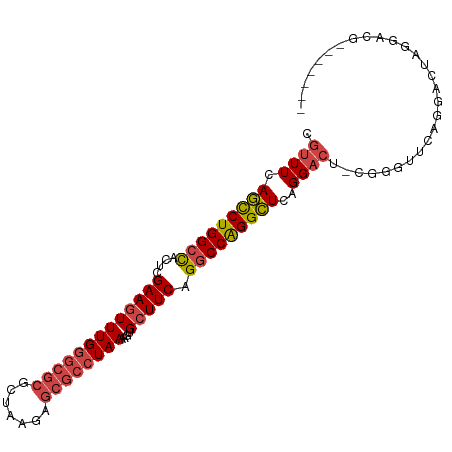
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:03 2011