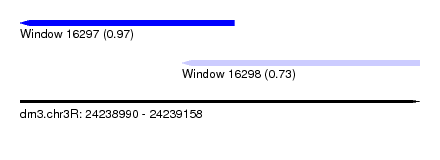
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,238,990 – 24,239,158 |
| Length | 168 |
| Max. P | 0.967181 |

| Location | 24,238,990 – 24,239,080 |
|---|---|
| Length | 90 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 70.76 |
| Shannon entropy | 0.56010 |
| G+C content | 0.44839 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -9.27 |
| Energy contribution | -12.01 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24238990 90 - 27905053 ----------UUCUUUUUUUUUCCACCCAUUUCGGCCCUUUUAUCCUCCGCCAGCGCUCCUUGCAGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA ----------....................((((((..((((((..((((((.(((((......))))))))))).))))))..)))))).......... ( -23.60, z-score = -3.36, R) >droSim1.chr3R 23926389 93 - 27517382 -------UUCUUUUUUUUUUUUCUACCGACUUCGGCCCUUUUAUCCUCCGCCAGCACUCCUUGCAGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA -------....................(((((((((..((((((..((((((.(((((......))))))))))).))))))..))))))..)))..... ( -27.00, z-score = -4.55, R) >droSec1.super_22 842374 90 - 1066717 ----------UUCUUUUUUUUUCUACCGACUUCGGCCCUUUUAUCCUCCGCCAGCACUCCUUGCGGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA ----------.................(((((((((..((((((..((((((.(((((......))))))))))).))))))..))))))..)))..... ( -27.00, z-score = -4.08, R) >droYak2.chr3R 6571600 100 + 28832112 UUCCUUUUUUUCCACCCCCCCACCUCCCAUUUUGGCCCUUUUAUCCUCCGCGAACGCUCCUUGCAGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA ..............................((..((..((((((((.(((((...((.....))..))))).)))..)))))..))..)).......... ( -19.30, z-score = -1.95, R) >droEre2.scaffold_4820 6667121 87 + 10470090 -----------UUCUUUCCCCCCCACC-AUUUCGGCACUUUUAUCCUCCGCCAG-GCUCUUUGCAGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA -----------................-..(((((((.((((((((.((((...-((.....))...)))).)))..))))).))))))).......... ( -22.60, z-score = -2.63, R) >droAna3.scaffold_13340 17039329 93 - 23697760 -------CUCCUACUCUUGCCUGAGGAUCCUUCCGCUUCCCCCGGCCACCCCGGUUCGCCUUGCAGUGUGGUUGAAAUAAAAAUGUUGAAUCGUCAUAAA -------(..((((.((.((..(.(((....)))((.....((((.....))))...)))..)))).))))..).......................... ( -15.10, z-score = 1.01, R) >droWil1.scaffold_181089 8904479 91 - 12369635 ---------UCCCCAUCCACCAGCAACAUUCUGGAUUCUCUUACACUAAGCGCAGAUGCCUUGCAUUGCGGUUGAAAUAAAAAUGUUGAAUCGUCAUAAA ---------..............(((((((..((.....)).....(((.(((((.(((...)))))))).))).......)))))))............ ( -13.50, z-score = 0.60, R) >consensus __________UUCCUUUUUCCUCCACCCAUUUCGGCCCUUUUAUCCUCCGCCAGCACUCCUUGCAGUGCGGUGGAAAUAAAAAUGUUGAAUCGUCAUAAA ..............................((((((..((((((..((((((.(((((......))))))))))).))))))..)))))).......... ( -9.27 = -12.01 + 2.74)


| Location | 24,239,058 – 24,239,158 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.59 |
| Shannon entropy | 0.59982 |
| G+C content | 0.45059 |
| Mean single sequence MFE | -20.05 |
| Consensus MFE | -9.38 |
| Energy contribution | -9.90 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24239058 100 - 27905053 UUUAUUUACC--CACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUAGCUACUUCCUGUUUUCU---UUUUUUUUCCACCCAUUU---------- ..........--.......(((.(((..(((......((((((((.......))))))))......)))..)))))).......---..................---------- ( -20.90, z-score = -1.45, R) >droSim1.chr3R 23926457 103 - 27517382 UUUAUUUACC--CACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCUUUUUUUUUUUUCUACCGACUU---------- ........((--(.....)))(((((.(((.......((((((((.......))))))))))).)))))....................................---------- ( -21.81, z-score = -1.69, R) >droSec1.super_22 842442 100 - 1066717 UUUAUUUACC--CACACCGGGGCAAACGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCU---UUUUUUUUCUACCGACUU---------- ........((--(.....)))((((..(((.......((((((((.......)))))))))))..))))...............---..................---------- ( -18.21, z-score = -0.89, R) >droYak2.chr3R 6571668 110 + 28832112 UUUAUUUACC--CACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGCUGCUGCUCCUUGCUACUUCCUGUUUUCCUUUUUUUCCACCCCCCCACCUCCCAUUU--- ........((--(.....)))(((((.(((.......((((((((.......))))))))))).)))))...........................................--- ( -21.81, z-score = -1.72, R) >droEre2.scaffold_4820 6667188 98 + 10470090 UUUAUUUACC--CACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUCCUUGCUACUUCCUGUUUUCUUUCCCCCCCACCAUUU--------------- ........((--(.....)))(((((.(((.(((((.....)))))....(((....)))))).)))))...............................--------------- ( -16.80, z-score = 0.02, R) >droAna3.scaffold_13340 17039387 112 - 23697760 UUUAUUUACC--CACACCGGAGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGAUGAUUCUGCUUC-GGCUCCUGCUUUUGCUCCUACUCUUGCCUGAGGAUCCUUCCGCUUCCCC ..........--......(((((((((((.......((((((..((((......)))))))))).-.....)))))..)))))).......((..(((....)))..))...... ( -25.42, z-score = -0.81, R) >dp4.chr2 20499326 80 + 30794189 UUUAUUUACCCACACACCGGAGCAAGCGGCUUUUACCAGCAGUAAAUUUAAGUUGAUGCUUGCUC-GAAU-CUCCCCCUGCC--------------------------------- ...................(((((((((((((((((.....))))....))))...)))))))))-....-...........--------------------------------- ( -16.00, z-score = -1.12, R) >droPer1.super_0 10186814 80 - 11822988 UUUAUUUACCCACACACCGGAGCAAGCGGCUUUUACCAGCAGUAAAUUUAAGUUGAUGCUUGCUC-GAAU-CUCCCCCCGCC--------------------------------- ...................(((((((((((((((((.....))))....))))...)))))))))-....-...........--------------------------------- ( -16.00, z-score = -1.35, R) >droWil1.scaffold_181089 8904548 100 - 12369635 UUUAUUUGCC--CACACCGGAGCUGGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUUCCGCACAGUUUCCGCUUCCCCA-UCCACCAGCAACAUUC------------ ..........--.....((((((((((((.......((((((((............)))))))))))).))).)))))........-................------------ ( -23.51, z-score = -1.50, R) >consensus UUUAUUUACC__CACACCGGGGCAAGCGGCUUUUACAAGCAGUAAAUUUAAGUUGAUGCUGCUCCUGGCUACUUCCUGUUUUCU___UUUUCCAUCAACC_AC_U__________ ..................((((((..((((((((((.....))))....))))))....)))))).................................................. ( -9.38 = -9.90 + 0.52)

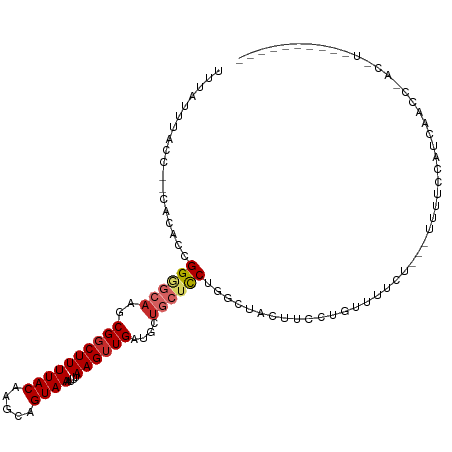
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:51:00 2011