
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,224,766 – 24,224,866 |
| Length | 100 |
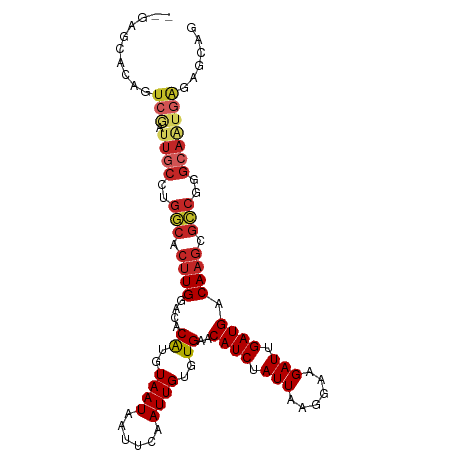
| Max. P | 0.967954 |

| Location | 24,224,766 – 24,224,866 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 85.23 |
| Shannon entropy | 0.32478 |
| G+C content | 0.44481 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -18.35 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.902273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
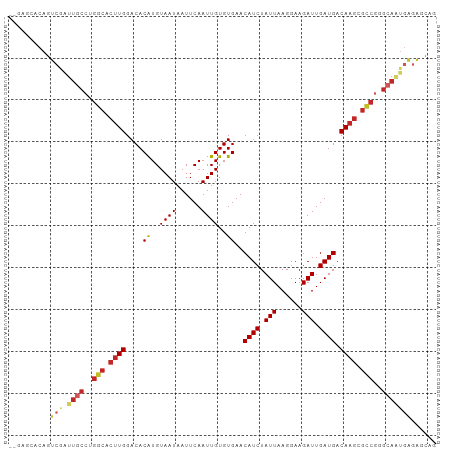
>dm3.chr3R 24224766 100 + 27905053 --CAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG --..((.(..((.((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))))).)).. ( -33.50, z-score = -3.57, R) >droAna3.scaffold_13340 17025160 100 + 23697760 --GACCCGAGUCGAUUGCCUGGCACUUGGACACGUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUAAGAGCAA --(((....))).((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))........ ( -32.80, z-score = -3.42, R) >droEre2.scaffold_4820 6652381 100 - 10470090 --CAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG --..((.(..((.((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))))).)).. ( -33.50, z-score = -3.57, R) >droYak2.chr3R 6556622 100 - 28832112 --GAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG --..((.(..((.((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))))).)).. ( -33.50, z-score = -3.63, R) >droSec1.super_22 828093 100 + 1066717 --GAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG --..((.(..((.((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))))).)).. ( -33.50, z-score = -3.63, R) >droSim1.chr3R 23911634 100 + 27517382 --GAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG --..((.(..((.((((((((((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))))))))))))).)).. ( -33.50, z-score = -3.63, R) >droMoj3.scaffold_6540 16965589 98 + 34148556 ---CAACCAGUCAGUUGCCAGCCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACA-GCGGCAAGCAGUGGGGCCAG ---......(((..((((..(((.((.(....((..((((......))))..))..((((.(((......))).)))).))-).)))..))))...)))... ( -20.20, z-score = 1.11, R) >droVir3.scaffold_12822 3651138 99 + 4096053 ---CAACCAGUCAGUUGCCAGCCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGGCGGGCAGUGAGGUCCG ---..(((..(((.(((((.(((.((((....((..((((......))))..))..((((.(((......))).)))).)))).))).)))))))))))... ( -30.10, z-score = -2.14, R) >droGri2.scaffold_14906 1257964 99 + 14172833 CAGCAAGCAGUCAGUUGCCAGCCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCAGCGAGAAUGGAAGC--- ......((..(((....(..((..((((....((..((((......))))..))..((((.(((......))).)))).))))..))..)..)))..))--- ( -16.90, z-score = 1.29, R) >droWil1.scaffold_181089 8887838 80 + 12369635 ---------------------GCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAA-CGGGUGGUAGAAGGAGCAG ---------------------.((((((....((..((((......))))..))..((((.(((......))).))))....-))))))............. ( -14.50, z-score = -0.40, R) >consensus __GAGCACAGUCGAUUGCCUGGCACUUGGACACAUGUAAUAAUUCAAUUGUGUGAACAUCUAUUAAGGAAGAUUGAUGACAAGCGCCGGGCAAUGAGAGCAG .............(((((..(((.((((....((..((((......))))..))..((((.(((......))).)))).)))).)))..)))))........ (-18.35 = -18.96 + 0.61)

| Location | 24,224,766 – 24,224,866 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.23 |
| Shannon entropy | 0.32478 |
| G+C content | 0.44481 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
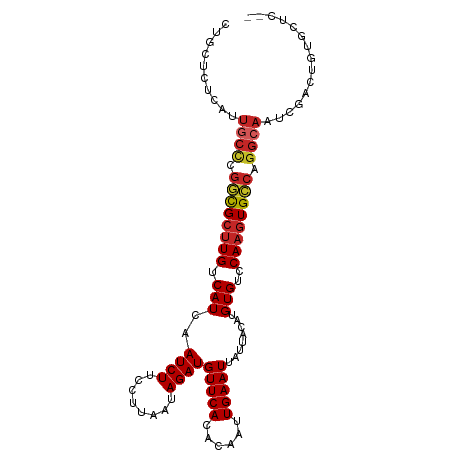
| SVM RNA-class probability | 0.967954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
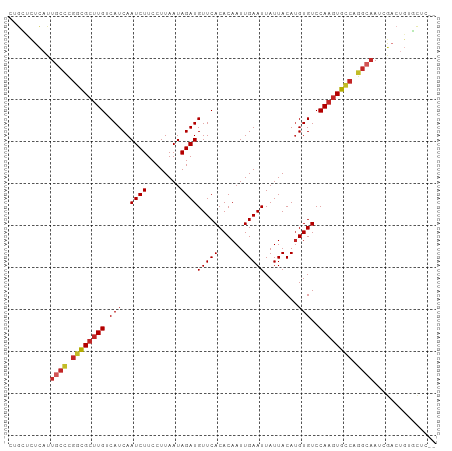
>dm3.chr3R 24224766 100 - 27905053 CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUG-- ..((..((((((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).)))))).))..))....-- ( -28.80, z-score = -3.08, R) >droAna3.scaffold_13340 17025160 100 - 23697760 UUGCUCUUAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACACGUGUCCAAGUGCCAGGCAAUCGACUCGGGUC-- ..((((..((((((.((((((((.(((..((((........))))(((((......)))))........)))..)))))))).))))))......)))).-- ( -26.80, z-score = -2.44, R) >droEre2.scaffold_4820 6652381 100 + 10470090 CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUG-- ..((..((((((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).)))))).))..))....-- ( -28.80, z-score = -3.08, R) >droYak2.chr3R 6556622 100 + 28832112 CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUC-- ..((..((((((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).)))))).))..))....-- ( -28.80, z-score = -3.23, R) >droSec1.super_22 828093 100 - 1066717 CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUC-- ..((..((((((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).)))))).))..))....-- ( -28.80, z-score = -3.23, R) >droSim1.chr3R 23911634 100 - 27517382 CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUC-- ..((..((((((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).)))))).))..))....-- ( -28.80, z-score = -3.23, R) >droMoj3.scaffold_6540 16965589 98 - 34148556 CUGGCCCCACUGCUUGCCGC-UGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGGCUGGCAACUGACUGGUUG--- ..((((.((.((((.(((((-(.......((((........))))....(((((..(((....)))..)))))...)))))).))))..))...)))).--- ( -21.20, z-score = 0.12, R) >droVir3.scaffold_12822 3651138 99 - 4096053 CGGACCUCACUGCCCGCCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGGCUGGCAACUGACUGGUUG--- ..(((((((.((((.((((((((......((((........))))....(((((..(((....)))..))))).)))))))).))))..)))..)))).--- ( -29.00, z-score = -2.75, R) >droGri2.scaffold_14906 1257964 99 - 14172833 ---GCUUCCAUUCUCGCUGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGGCUGGCAACUGACUGCUUGCUG ---((..........((..((((......((((........))))....(((((..(((....)))..))))).))))..)).((((......)))).)).. ( -18.10, z-score = -0.02, R) >droWil1.scaffold_181089 8887838 80 - 12369635 CUGCUCCUUCUACCACCCG-UUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGC--------------------- .............(((...-.........((((........))))....(((((..(((....)))..)))))....))).--------------------- ( -5.10, z-score = 1.45, R) >consensus CUGCUCUCAUUGCCCGGCGCUUGUCAUCAAUCUUCCUUAAUAGAUGUUCACACAAUUGAAUUAUUACAUGUGUCCAAGUGCCAGGCAAUCGACUGUGCUC__ ..........((((.((((((((.(((..((((........))))(((((......)))))........)))..)))))))).))))............... (-20.23 = -20.54 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:58 2011