| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,220,479 – 24,220,575 |
| Length | 96 |
| Max. P | 0.678455 |

| Location | 24,220,479 – 24,220,575 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 85.64 |
| Shannon entropy | 0.30301 |
| G+C content | 0.53479 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -22.62 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678455 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
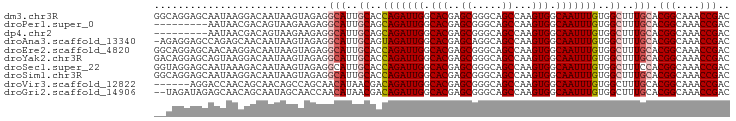
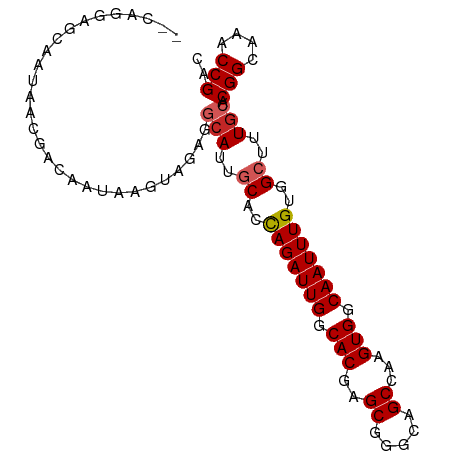
>dm3.chr3R 24220479 96 - 27905053 GGCAGGAGCAAUAAGGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC .((....))....................(((..((..(((((((.(((..((.....))...))).)))))))..))..))).(((....))).. ( -27.40, z-score = -0.73, R) >droPer1.super_0 10167139 87 - 11822988 ---------AAUAACGACAGUAAGAAGAGGCAUUGCAGCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC ---------....................(((..((.((((((((.(((..((.....))...))).)))))))).))..))).(((....))).. ( -28.00, z-score = -1.99, R) >dp4.chr2 20479565 87 + 30794189 ---------AAUAACGACAGUAAGAAGAGGCAUUGCAGCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC ---------....................(((..((.((((((((.(((..((.....))...))).)))))))).))..))).(((....))).. ( -28.00, z-score = -1.99, R) >droAna3.scaffold_13340 17021030 95 - 23697760 -AGAGGAGCCAGAGCAACAAUAAGUAGAGGCAUUGCAGUAGAUUGGCACGAGCAGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC -...((.(((...((........))....(((..((..(((((((.(((..((.....))...))).)))))))..))..)))..)))...))... ( -27.10, z-score = -0.98, R) >droEre2.scaffold_4820 6648056 96 + 10470090 GGCAGGAGCAACAAGGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC .((....))....................(((..((..(((((((.(((..((.....))...))).)))))))..))..))).(((....))).. ( -27.40, z-score = -0.57, R) >droYak2.chr3R 6552131 96 + 28832112 GACAGGAGCAGUAAGGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC ....((....((....)).....((....(((..((..(((((((.(((..((.....))...))).)))))))..))..)))...))...))... ( -26.40, z-score = -0.66, R) >droSec1.super_22 823869 96 - 1066717 GGUAGGAGCAAUAAAGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUCCACGGCAAACCGAC (((.((.(((((....((.....))......))))).))((((((.(((..((.....))...))).))))))((((....)))).....)))... ( -25.20, z-score = -0.45, R) >droSim1.chr3R 23907382 96 - 27517382 GGCAGGAGCAAUAAGGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC .((....))....................(((..((..(((((((.(((..((.....))...))).)))))))..))..))).(((....))).. ( -27.40, z-score = -0.73, R) >droVir3.scaffold_12822 3640557 90 - 4096053 ------AGGACCAACAGCAACAGCCAGCAACAUAACGACAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC ------..........((((.((((.....(.....)((((((((.(((..((.....))...))).)))))))))))))))).(((....))).. ( -27.70, z-score = -1.56, R) >droGri2.scaffold_14906 1250031 94 - 14172833 --UAGAUAGAGCAACAGCAAUAGCAACCAACAUAACGACAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC --..(.((((((....((....)).............((((((((.(((..((.....))...))).)))))))).))))))).(((....))).. ( -26.20, z-score = -1.98, R) >consensus __CAGGAGCAAUAACGACAAUAAGUAGAGGCAUUGCACCAGAUUGGCACGAGCGGGCAGCCAAGUGGCAAUUUGUGGCUUUGCACGGCAAACCGAC .............................(((..((..(((((((.(((..((.....))...))).)))))))..))..))).(((....))).. (-22.62 = -23.03 + 0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:56 2011