
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,217,163 – 24,217,254 |
| Length | 91 |
| Max. P | 0.741790 |

| Location | 24,217,163 – 24,217,254 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Shannon entropy | 0.42371 |
| G+C content | 0.60647 |
| Mean single sequence MFE | -44.06 |
| Consensus MFE | -14.43 |
| Energy contribution | -14.12 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24217163 91 - 27905053 GAGGUAAGAUGGCGG-------GCGGAAGUG-GCAAGGGGCG-----------------GUCGCGGCGAACGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..((((((((((.((-------((((..(((-((..((((((-----------------.(((...))).))))))....(((---......))))))))..)))))).)))))))))) ( -48.30, z-score = -4.20, R) >droSim1.chr3R 23904042 100 - 27517382 GAGGUAAGAUGGCGG-------GCGGAAGUG-GCAUAGGGUGUGUGGGGCG--------GGCGCGGCGAACGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..((((((((((.((-------((((..(((-((((..(((((.(((((((--------..((...))..)))))))...)).---)))..))..)))))..)))))).)))))))))) ( -52.20, z-score = -3.82, R) >droSec1.super_22 820534 100 - 1066717 GAGGUAAGAUGGCGG-------GCGGAAGUG-GCAUAGGGUGUGUGGGGCG--------GGCGCGGCGAACGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..((((((((((.((-------((((..(((-((((..(((((.(((((((--------..((...))..)))))))...)).---)))..))..)))))..)))))).)))))))))) ( -52.20, z-score = -3.82, R) >droYak2.chr3R 6548779 115 + 28832112 GAGGUAAGAUGGCGGGUGGAAGGCGGAAGUG-GCAAGGAGUGUGCGGGGGGCGGGGCGUGGCACGGCGAACGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..((((((((((.(((..(...((....)).-.....(.(((..(.((..(((((((((.((...))..)))))))....)).---.))..)..))))....)..))).)))))))))) ( -51.70, z-score = -2.16, R) >droEre2.scaffold_4820 6644759 94 + 10470090 GAGGUAAGAUGGCGG-------GCGGAAGUG-GCAAGGAGUGUGC--------------GGCGCAGCUAACGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..((((((((((.((-------((((..(((-((.(((.((((..--------------(((...))).)))))))....(((---......))))))))..)))))).)))))))))) ( -44.50, z-score = -3.33, R) >droAna3.scaffold_13340 17017182 96 - 23697760 GAGGUAAGAUGGCGGCU-----GCGGAGCUAUGUGGGUGGUGGG---------------GGCGUGGCAAGCGCCCUAAUAGCA---ACCAGGUGCGCCAUAAUUGCCAAACAUUUUGGC ..(((((.(((((((((-----....)))(((.(((...((.((---------------(((((.....)))))))....)).---.))).)))))))))..)))))............ ( -37.80, z-score = -1.06, R) >droPer1.super_0 10164218 91 - 11822988 GAGGUAAGAUGGUGG-------GAGGACG---GCCCG-AGCA-----------------GGAGCAGGAGCCGCCCUAAUAGCAGCGACCAGGUGUGGCAUAAUUGCCAAACAUUUUCCU ..(((....((.((.-------.(((.((---((((.-.((.-----------------...)).)).)))).)))..)).))...)))..((.(((((....))))).))........ ( -32.90, z-score = -1.76, R) >dp4.chr2 20476646 91 + 30794189 GAGGUAAGAUGGUGG-------GAGGACG---GCCCG-AGCA-----------------GGAGCAGGAGCCGCCCUAAUAGCAGCGACCAGGUGUGGCAUAAUUGCCAAACAUUUUCCU ..(((....((.((.-------.(((.((---((((.-.((.-----------------...)).)).)))).)))..)).))...)))..((.(((((....))))).))........ ( -32.90, z-score = -1.76, R) >consensus GAGGUAAGAUGGCGG_______GCGGAAGUG_GCAAGGAGUGUG_______________GGCGCGGCGAACGCCCUAAUAGCA___ACCAGGUGCGCCAUAAUUGCCCACCGUUUUGCC ..(((((((((.....................((.....))..................((((.......((((((.............))).))).......))))...))))))))) (-14.43 = -14.12 + -0.31)

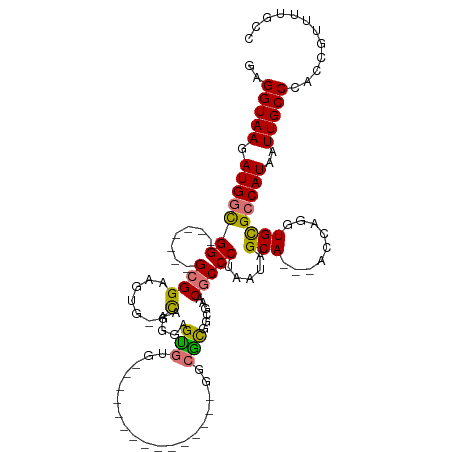
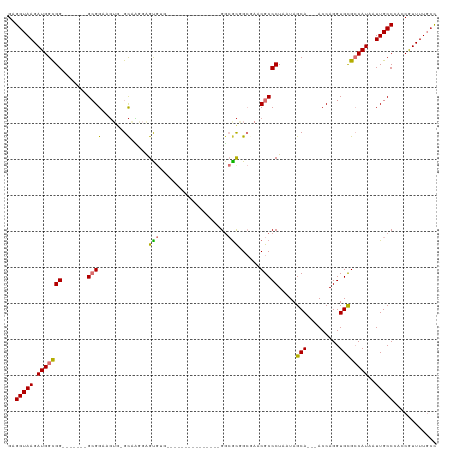
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:54 2011