
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,216,481 – 24,216,558 |
| Length | 77 |
| Max. P | 0.774618 |

| Location | 24,216,481 – 24,216,558 |
|---|---|
| Length | 77 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 85.45 |
| Shannon entropy | 0.31580 |
| G+C content | 0.54558 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
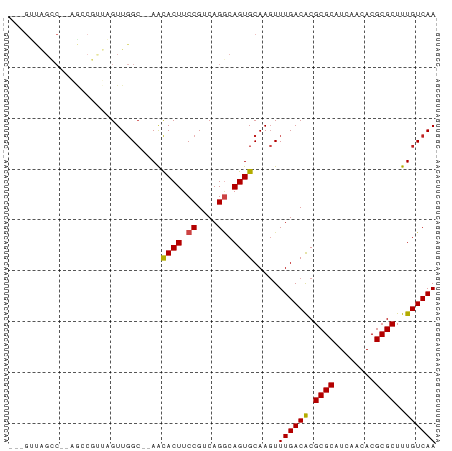
>dm3.chr3R 24216481 77 + 27905053 ---GUUAGCC--AGCCGUUAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---....(((--(((.....))))))--..((((.((....)).)))).....((((((.((((........))))..)))))) ( -26.80, z-score = -2.53, R) >droSim1.chr3R 23903367 77 + 27517382 ---GUUAGCC--AGCCGUUAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---....(((--(((.....))))))--..((((.((....)).)))).....((((((.((((........))))..)))))) ( -26.80, z-score = -2.53, R) >droSec1.super_22 819853 77 + 1066717 ---GUUAGCC--AGCCGUUAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---....(((--(((.....))))))--..((((.((....)).)))).....((((((.((((........))))..)))))) ( -26.80, z-score = -2.53, R) >droYak2.chr3R 6548081 77 - 28832112 ---GUUAGCC--AGCCGUUAGUUGGC--AACACUUCCGUCAGGCAGUGCAACUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---(((.(((--(((.....))))))--..((((.((....)).)))).))).((((((.((((........))))..)))))) ( -27.00, z-score = -2.74, R) >droEre2.scaffold_4820 6644068 77 - 10470090 ---GUUAGCC--AGCCGUAAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---....(((--(((.....))))))--..((((.((....)).)))).....((((((.((((........))))..)))))) ( -26.80, z-score = -2.46, R) >droAna3.scaffold_13340 17016477 76 + 23697760 ---GUUAGCC--CGCCG-GAGUUGGA--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---....(((--.(.((-(((.((..--..)).))))).).))).........((((((.((((........))))..)))))) ( -28.20, z-score = -2.45, R) >dp4.chr2 20475588 77 - 30794189 ---GUUGGCC--AAACGCUAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---((..(((--..(((..(((.(..--..))))..)))..)))...))....((((((.((((........))))..)))))) ( -25.10, z-score = -1.29, R) >droPer1.super_0 10163177 77 + 11822988 ---GUUGGCC--AAACGCUAGUUGGC--AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---((..(((--..(((..(((.(..--..))))..)))..)))...))....((((((.((((........))))..)))))) ( -25.10, z-score = -1.29, R) >droWil1.scaffold_181089 8875333 84 + 12369635 GGUGGCAGACUUAAGCCCUUGUUGGCGUAGUACUUCCGUCAGACAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA .(((.(((((((..(((......)))...(((((((.....)).)))))))))))).)))((((........))))........ ( -25.90, z-score = -0.69, R) >droGri2.scaffold_14906 1245831 76 + 14172833 ---GCUGGGC---GGACAUUGUUGGC--AGCACUUCCGGCAGUCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ---..((.((---.(((..((((((.--.......))))))))).)).))...((((((.((((........))))..)))))) ( -23.10, z-score = 0.35, R) >droMoj3.scaffold_6540 16950674 62 + 34148556 --------------------GUUCGC--AGCACUUCCGGCAGGCAGUGCAAGUUUGACGCGCGCGUCAACACGCGCUUUGUCAA --------------------......--.(((((.((....)).)))))....((((((.((((((....))))))..)))))) ( -24.50, z-score = -1.57, R) >droVir3.scaffold_12822 3635671 62 + 4096053 --------------------GUUGGC--AGCACUUCCGGCAGGCAGUGCAAGUUUGACGCGCGCAUCAACACGCGCUUUGUCAA --------------------.(((((--((((((.((....)).))))).........(((((........)))))..)))))) ( -23.80, z-score = -1.46, R) >consensus ___GUUAGCC__AGCCGUUAGUUGGC__AACACUUCCGUCAGGCAGUGCAAGUUUGACACGCGCAUCAACACGCGCUUUGUCAA ..............................((((.((....)).)))).....((((((.((((........))))..)))))) (-16.70 = -16.65 + -0.05)


| Location | 24,216,481 – 24,216,558 |
|---|---|
| Length | 77 |
| Sequences | 12 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 85.45 |
| Shannon entropy | 0.31580 |
| G+C content | 0.54558 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -20.63 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24216481 77 - 27905053 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUAACGGCU--GGCUAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--((((.(....)..)--)))....--- ( -27.70, z-score = -1.99, R) >droSim1.chr3R 23903367 77 - 27517382 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUAACGGCU--GGCUAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--((((.(....)..)--)))....--- ( -27.70, z-score = -1.99, R) >droSec1.super_22 819853 77 - 1066717 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUAACGGCU--GGCUAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--((((.(....)..)--)))....--- ( -27.70, z-score = -1.99, R) >droYak2.chr3R 6548081 77 + 28832112 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAAGUUGCACUGCCUGACGGAAGUGUU--GCCAACUAACGGCU--GGCUAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--((((.(....)..)--)))....--- ( -27.70, z-score = -1.60, R) >droEre2.scaffold_4820 6644068 77 + 10470090 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUUACGGCU--GGCUAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--((((.(.....).)--)))....--- ( -26.40, z-score = -1.44, R) >droAna3.scaffold_13340 17016477 76 - 23697760 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--UCCAACUC-CGGCG--GGCUAAC--- ((((((..((((((....)))))).)))))).........(((((.((((..((..--..))..))-)).))--)))....--- ( -29.80, z-score = -2.45, R) >dp4.chr2 20475588 77 + 30794189 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUAGCGUUU--GGCCAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--(((((.......))--)))....--- ( -26.10, z-score = -0.97, R) >droPer1.super_0 10163177 77 - 11822988 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU--GCCAACUAGCGUUU--GGCCAAC--- ((((((..((((((....)))))).))))))....(((((.((....)).))))).--(((((.......))--)))....--- ( -26.10, z-score = -0.97, R) >droWil1.scaffold_181089 8875333 84 - 12369635 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGUCUGACGGAAGUACUACGCCAACAAGGGCUUAAGUCUGCCACC ((((((..((((((....)))))).)))))).((((...(((...((....))...)))....))))(((........)))... ( -26.00, z-score = -0.82, R) >droGri2.scaffold_14906 1245831 76 - 14172833 UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGACUGCCGGAAGUGCU--GCCAACAAUGUCC---GCCCAGC--- ((((((..((((((....)))))).))))))....(((((..(....)..))))).--.............---.......--- ( -22.30, z-score = -0.04, R) >droMoj3.scaffold_6540 16950674 62 - 34148556 UUGACAAAGCGCGUGUUGACGCGCGCGUCAAACUUGCACUGCCUGCCGGAAGUGCU--GCGAAC-------------------- (((((...(((((((....))))))))))))....(((((.((....)).))))).--......-------------------- ( -25.50, z-score = -1.92, R) >droVir3.scaffold_12822 3635671 62 - 4096053 UUGACAAAGCGCGUGUUGAUGCGCGCGUCAAACUUGCACUGCCUGCCGGAAGUGCU--GCCAAC-------------------- (((((...((((((......)))))))))))....(((((.((....)).))))).--......-------------------- ( -23.10, z-score = -1.51, R) >consensus UUGACAAAGCGCGUGUUGAUGCGCGUGUCAAACUUGCACUGCCUGACGGAAGUGUU__GCCAACUAACGGCU__GGCUAAC___ (((((...((((((....))))))..)))))....(((((.((....)).)))))............................. (-20.63 = -20.43 + -0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:54 2011