| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,876,054 – 8,876,169 |
| Length | 115 |
| Max. P | 0.931742 |

| Location | 8,876,054 – 8,876,169 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.61 |
| Shannon entropy | 0.65850 |
| G+C content | 0.51637 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.84 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
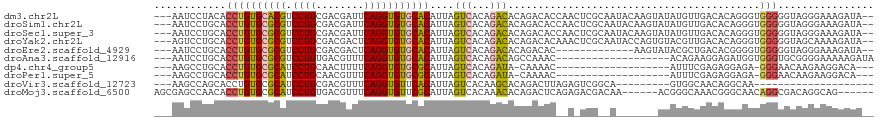
>dm3.chr2L 8876054 115 + 23011544 ---AAUCCUACACCUGUGCACGUCCUGCGACGAUUCAGGUGUGCACAUUAGUCACAGACACAGACACCAACUCGCAAUACAAGUAUAUGUUGACACAGGGUGGGGGUAGGGAAAGAUA-- ---..((((((.((((((((((.((((........)))))))))))....(((...))).....((((......(((((........)))))......))))))))))))).......-- ( -37.30, z-score = -1.99, R) >droSim1.chr2L 8654894 115 + 22036055 ---AAUCCUGCACCUGUGCGCGUCCUGCGACGAUUCAGGUGUGCACAUUAGUCACAGACACAGACACCAACUCGCAAUACAAGUAUAUGUUGACACAGGGUGGGGGUAGGGAAAGAUA-- ---..((((((.((((((((((.((((........)))))))))))....(((...))).....((((......(((((........)))))......))))))))))))).......-- ( -36.90, z-score = -1.25, R) >droSec1.super_3 4350084 115 + 7220098 ---AAUCCUGCACCUGUGCGCGUCCUGCGACGAUUCAGGUGUGCACAUUAGUCACAGACACAGACACCAACUCGCAAUACAAGUAUAUGUUGACACAGGGUGGGGGUAGGGAAAGAUA-- ---..((((((.((((((((((.((((........)))))))))))....(((...))).....((((......(((((........)))))......))))))))))))).......-- ( -36.90, z-score = -1.25, R) >droYak2.chr2L 11523650 115 + 22324452 ---AGUCCUGCACCUGUGCGCGUCCUGCGACGACUCAGGUGUGCACAUUAGUCACAGACACAGACACAAACUCGCAAUACCAGUGUACGUUGACACAGGGUGGGGGUAGCAAAAGAUA-- ---.(((((.((((((((((((.((((.(....).)))))))))))....(((((..((((.....................))))..).))))...))))).)))..))........-- ( -35.90, z-score = -0.07, R) >droEre2.scaffold_4929 9469275 102 + 26641161 ---AAUCCUGCACCUGUGCGCGUCCUGCGACGACUCAGGUGUGCACAUUAGUCACAGACACAGACAC-------------AAGUAUACGCUGACACGGGGUGGGGGUAGGGAAAGAUA-- ---..((((((.((((((((((.((((.(....).)))))))))))....(((.........)))..-------------.....(((.(((...))).)))))))))))).......-- ( -32.80, z-score = -0.42, R) >droAna3.scaffold_12916 10540639 98 + 16180835 ---AAUCCUGCACCUGUGCGCGUCCUGUGACGUUUCAGGUGUGCACAUUAGUCACAGACAGCCAAAC-------------------ACAGAAGGAGAUGGUGGGUGCGGGGAAAAAGAUA ---..(((((((((((((((((.((((........)))))))))))....(((...))).((((...-------------------.(....)....))))))))))))))......... ( -37.40, z-score = -2.55, R) >dp4.chr4_group5 1684206 93 - 2436548 ---AAGCCUGCACCUGUGCGCAUCCUGCAACUUUUCAGGUGUGCGCAUUAGUCACAGAUA-CAAAAC-------------------AUUUCGAGAGGAGA-GGGAACAAGAAGGACA--- ---..((((...(((((((((((((((........)))).))))))))............-......-------------------.((((....)))))-))........))).).--- ( -23.40, z-score = -0.23, R) >droPer1.super_5 1655245 93 - 6813705 ---AAGCCUGCACCUGUGCGCAUCCUGCAACGUUUCAGGUGUGCGCAUUAGUCACAGAUA-CAAAAC-------------------AUUUCGAGAGGAGA-GGGAACAAGAAGGACA--- ---..((((...(((((((((((((((........)))).))))))))............-......-------------------.((((....)))))-))........))).).--- ( -23.40, z-score = -0.11, R) >droVir3.scaffold_12723 4896317 88 + 5802038 ---AAGCCAGCACCUGUGCGCAUCCUGCGACGUUUCAGGUGUUCACAUUAGUCACAAGCACAGACUUAGAGUCGGCA---------GUGGCAACAGGCAA-------------------- ---..(((((((((((..(((.....)))......)))))))).......(((((..((...(((.....))).)).---------)))))....)))..-------------------- ( -31.60, z-score = -2.55, R) >droMoj3.scaffold_6500 10884131 108 + 32352404 AGCGAGCCAACACCUGUGCGCAUCCUGUGACGUUUCAGGUGUUCGCAUUAGUCACAAACACAGACUCAGAGACGACAA------ACGGGCAAACGGGCAACAGGCGACAGGCAG------ .....(((..(.(((((((.(..(((((..((((((.((((....))))((((.........))))..))))))....------))))).....).)).))))).)...)))..------ ( -31.40, z-score = -0.63, R) >consensus ___AAUCCUGCACCUGUGCGCGUCCUGCGACGAUUCAGGUGUGCACAUUAGUCACAGACACAGACAC__A_UCG__A_______AUAUGUUGACACAGGGUGGGGGUAGGGAAAGAUA__ ............((((((((((.((((........)))))))))))....(((...)))..........................................)))................ (-17.01 = -16.84 + -0.17)

| Location | 8,876,054 – 8,876,169 |
|---|---|
| Length | 115 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.61 |
| Shannon entropy | 0.65850 |
| G+C content | 0.51637 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -16.13 |
| Energy contribution | -15.82 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.35 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.547333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
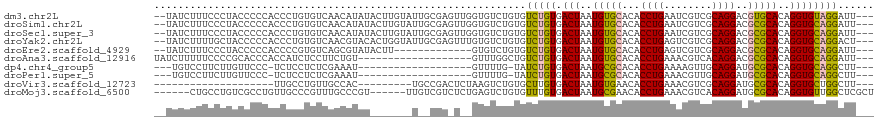
>dm3.chr2L 8876054 115 - 23011544 --UAUCUUUCCCUACCCCCACCCUGUGUCAACAUAUACUUGUAUUGCGAGUUGGUGUCUGUGUCUGUGACUAAUGUGCACACCUGAAUCGUCGCAGGACGUGCACAGGUGUAGGAUU--- --........(((((((.(((...(..(..((((..(((((.....)))))..))))..)..)..))).....(((((((.((((........))))..))))))))).)))))...--- ( -34.60, z-score = -1.26, R) >droSim1.chr2L 8654894 115 - 22036055 --UAUCUUUCCCUACCCCCACCCUGUGUCAACAUAUACUUGUAUUGCGAGUUGGUGUCUGUGUCUGUGACUAAUGUGCACACCUGAAUCGUCGCAGGACGCGCACAGGUGCAGGAUU--- --............((..((((.(((((..((((..(((((.....)))))..))))..(((((((((((...................)))))).))))))))))))))..))...--- ( -33.81, z-score = -0.80, R) >droSec1.super_3 4350084 115 - 7220098 --UAUCUUUCCCUACCCCCACCCUGUGUCAACAUAUACUUGUAUUGCGAGUUGGUGUCUGUGUCUGUGACUAAUGUGCACACCUGAAUCGUCGCAGGACGCGCACAGGUGCAGGAUU--- --............((..((((.(((((..((((..(((((.....)))))..))))..(((((((((((...................)))))).))))))))))))))..))...--- ( -33.81, z-score = -0.80, R) >droYak2.chr2L 11523650 115 - 22324452 --UAUCUUUUGCUACCCCCACCCUGUGUCAACGUACACUGGUAUUGCGAGUUUGUGUCUGUGUCUGUGACUAAUGUGCACACCUGAGUCGUCGCAGGACGCGCACAGGUGCAGGACU--- --....((((((.(((...(((..((((......)))).)))..........(((((..(((((((((((...................)))))).)))))))))))))))))))..--- ( -31.71, z-score = 1.21, R) >droEre2.scaffold_4929 9469275 102 - 26641161 --UAUCUUUCCCUACCCCCACCCCGUGUCAGCGUAUACUU-------------GUGUCUGUGUCUGUGACUAAUGUGCACACCUGAGUCGUCGCAGGACGCGCACAGGUGCAGGAUU--- --.....................(((....)))....(((-------------(..(((((((.((((((....)).))))((((.(....).))))....)))))))..))))...--- ( -27.50, z-score = 0.23, R) >droAna3.scaffold_12916 10540639 98 - 16180835 UAUCUUUUUCCCCGCACCCACCAUCUCCUUCUGU-------------------GUUUGGCUGUCUGUGACUAAUGUGCACACCUGAAACGUCACAGGACGCGCACAGGUGCAGGAUU--- ..........((.(((((.............(((-------------------((.((....((((((((...................)))))))).)).)))))))))).))...--- ( -25.22, z-score = -0.33, R) >dp4.chr4_group5 1684206 93 + 2436548 ---UGUCCUUCUUGUUCCC-UCUCCUCUCGAAAU-------------------GUUUUG-UAUCUGUGACUAAUGCGCACACCUGAAAAGUUGCAGGAUGCGCACAGGUGCAGGCUU--- ---................-..............-------------------..((((-(((((((.......(((((..((((........)))).))))))))))))))))...--- ( -22.71, z-score = -0.29, R) >droPer1.super_5 1655245 93 + 6813705 ---UGUCCUUCUUGUUCCC-UCUCCUCUCGAAAU-------------------GUUUUG-UAUCUGUGACUAAUGCGCACACCUGAAACGUUGCAGGAUGCGCACAGGUGCAGGCUU--- ---................-..............-------------------..((((-(((((((.......(((((..((((........)))).))))))))))))))))...--- ( -22.71, z-score = -0.29, R) >droVir3.scaffold_12723 4896317 88 - 5802038 --------------------UUGCCUGUUGCCAC---------UGCCGACUCUAAGUCUGUGCUUGUGACUAAUGUGAACACCUGAAACGUCGCAGGAUGCGCACAGGUGCUGGCUU--- --------------------.........((((.---------.((((((.....)))(((((.(((.((....))..)))((((........))))....))))))))..))))..--- ( -26.10, z-score = -0.51, R) >droMoj3.scaffold_6500 10884131 108 - 32352404 ------CUGCCUGUCGCCUGUUGCCCGUUUGCCCGU------UUGUCGUCUCUGAGUCUGUGUUUGUGACUAAUGCGAACACCUGAAACGUCACAGGAUGCGCACAGGUGUUGGCUCGCU ------..(((...((((((((((..((((((.((.------....))......((((.........))))...)))))).((((........))))..))).)))))))..)))..... ( -33.20, z-score = -0.63, R) >consensus __UAUCUUUCCCUACCCCCACCCCCUGUCAACAUAU_______U__CGA_U__GUGUCUGUGUCUGUGACUAAUGUGCACACCUGAAACGUCGCAGGACGCGCACAGGUGCAGGAUU___ ..............................................................(((((.(((..(((((...((((........))))..)))))..))))))))...... (-16.13 = -15.82 + -0.31)
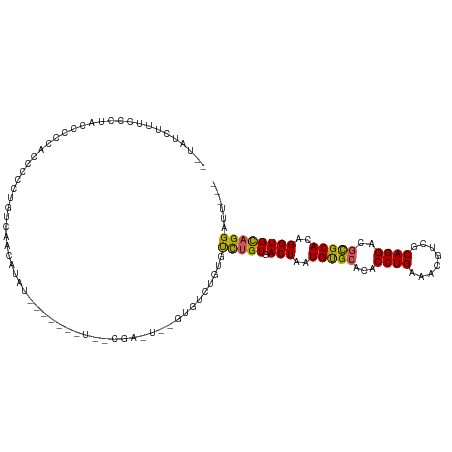
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:13 2011