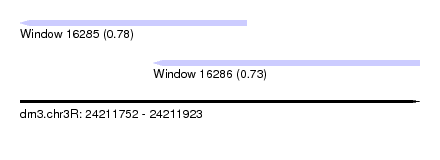
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,211,752 – 24,211,923 |
| Length | 171 |
| Max. P | 0.781043 |

| Location | 24,211,752 – 24,211,849 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 97.58 |
| Shannon entropy | 0.05016 |
| G+C content | 0.45378 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -28.87 |
| Energy contribution | -28.44 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24211752 97 - 27905053 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -29.60, z-score = -1.17, R) >droAna3.scaffold_13340 17012491 97 - 23697760 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -30.30, z-score = -1.53, R) >droEre2.scaffold_4820 6639271 97 + 10470090 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -29.60, z-score = -1.17, R) >droYak2.chr3R 6543362 97 + 28832112 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -29.60, z-score = -1.17, R) >droSec1.super_22 815112 97 - 1066717 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -29.60, z-score = -1.17, R) >droSim1.chr3R 23898573 97 - 27517382 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -29.60, z-score = -1.17, R) >droPer1.super_0 10159180 99 - 11822988 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAUUGCCAACAGAGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).)))....... ( -30.30, z-score = -1.32, R) >dp4.chr2 20471594 99 + 30794189 UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAACAGAGCCAG .....(((((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).)))).)))....... ( -30.30, z-score = -1.30, R) >droVir3.scaffold_12822 3630520 97 - 4096053 UAUUAUGUGGUGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....((((((..(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....)))..))).))--)..... ( -28.60, z-score = -1.05, R) >droMoj3.scaffold_6540 16946850 97 - 34148556 UAUUAUGUGGUACUGCUAUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....(((((((.(((..((((.......))))...((((.((((((.....((((....)))).)))))).))))....))).)))).))--)..... ( -25.90, z-score = -1.01, R) >droGri2.scaffold_14906 1241781 97 - 14172833 UAUUAUGUGGUGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUAUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC--AGCCAG .....((((((..(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....)))..))).))--)..... ( -28.60, z-score = -1.04, R) >consensus UAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCAGCAGGAAGAAUCAAGUAGUUUUUUAUGUUUUCCGGCUGAACUGCAAUGCCAAC__AGCCAG .......(((((.(((.(((((.......)))))..((((.((((((.....((((....)))).)))))).))))....))).))))).......... (-28.87 = -28.44 + -0.44)



| Location | 24,211,809 – 24,211,923 |
|---|---|
| Length | 114 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Shannon entropy | 0.39672 |
| G+C content | 0.50052 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24211809 114 - 27905053 ACUUUAAGUGCCAUAUUUCCUGCUGCUGCUCCA---ACUACCACUGA---GCUCCUCUGCCCCCCGCAGUUCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC ......(((((((........((.((.((.(((---.........((---((....((((.....))))...))))...........))).)).))...)).......)))))))..... ( -30.11, z-score = -2.00, R) >droAna3.scaffold_13340 17012548 97 - 23697760 ACUUUAAGUGCCAUAUUUCCU------GCUCCUG--AUUCCCCGUA---------------CCCCGUCGUCUGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCAGC .........(((((((.....------((...((--((........---------------....))))...))..........)))))))((((..(((((.......)))))..)))) ( -21.56, z-score = -2.28, R) >droEre2.scaffold_4820 6639328 111 + 10470090 ACUUUAAGUGCCAUAUUUCCUGCU---GCUCCA---ACUACCACUGA---GCUCCUCGGCCCCCCGCAGUCCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC .........(((((((...((((.---((((..---.........))---)).....((....))))))...............)))))))((((..(((((.......)))))..)))) ( -27.67, z-score = -0.99, R) >droYak2.chr3R 6543419 120 + 28832112 ACUUUAAGUGCCAUAUUUCCUGCUGCCGCUCCAACUACUACCACUGAGCUGCUCCUCGGCCCCCCGCAGUCCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC .........(((((((...((((.((.((((..............)))).)).....((....))))))...............)))))))((((..(((((.......)))))..)))) ( -30.41, z-score = -0.83, R) >droSec1.super_22 815169 114 - 1066717 ACUUUAAGUGCCAUAUUUCCUGCUGCUGCUCCA---ACUACCACUGA---GCUCCUCUGCCCCCCGCAGUCCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC ......(((((((........((.((.((.(((---.........((---((....((((.....))))...))))...........))).)).))...)).......)))))))..... ( -30.11, z-score = -2.00, R) >droSim1.chr3R 23898630 114 - 27517382 ACUUUAAGUGCCAUAUUUCCUGCUGCUGCUCCA---ACUACCACUGA---GCUCCUCUGCCCCCCGCAGUCCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC ......(((((((........((.((.((.(((---.........((---((....((((.....))))...))))...........))).)).))...)).......)))))))..... ( -30.11, z-score = -2.00, R) >droWil1.scaffold_181089 3499360 99 + 12369635 ACUUUAAGUGCCAUAUUUCCU------GCUCUUGCCAUGGCUCCUA---------------CUGUGUCGGCUGCUUUAUUUAUUAUGUGGCGCUACUGUGCUUACUUUUGGCACUCCAGC ......((((((((((.....------((..(((.(((((......---------------))))).)))..))..........))))))))))...(((((.......)))))...... ( -24.96, z-score = -1.38, R) >droVir3.scaffold_12822 3630577 87 - 4096053 ACUUUAAGUGCCAUAUU------------UCCU---GAUGCCGCUG------------------CGCUGCCUGCUUUAUUUAUUAUGUGGUGCUGCUGUGCUUACUUUUGGCACUCCAGC .........(((((((.------------....---((((..((.(------------------(...))..))..))))....)))))))((((..(((((.......)))))..)))) ( -22.30, z-score = -1.09, R) >droMoj3.scaffold_6540 16946907 93 - 34148556 ACUUUAAGUGCCAUAUUUCCU------GCUCCU---GCUGCUGCUG------------------CAUUGCCUGCUUUAUUUAUUAUGUGGUACUGCUAUGCUUACUUUUGGCACUCCAGC ......(((((((........------((....---))....((.(------------------((.((((.((............)))))).)))...)).......)))))))..... ( -21.10, z-score = -1.03, R) >consensus ACUUUAAGUGCCAUAUUUCCUGCU___GCUCCA___ACUACCACUGA___GCUCCUC_GCCCCCCGCAGUCCGCUUUAUUUAUUAUGUGGCGCUGCUGUGCUUACUUUUGGCACUCCGGC ......((((((((((...........((...........................................))..........))))))))))((((((((.......)))))...))) (-16.90 = -16.83 + -0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:51 2011