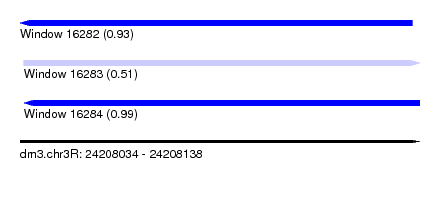
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,208,034 – 24,208,138 |
| Length | 104 |
| Max. P | 0.991461 |

| Location | 24,208,034 – 24,208,136 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.84 |
| Shannon entropy | 0.30753 |
| G+C content | 0.43468 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.925904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
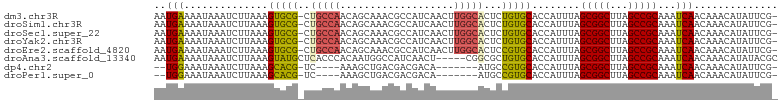
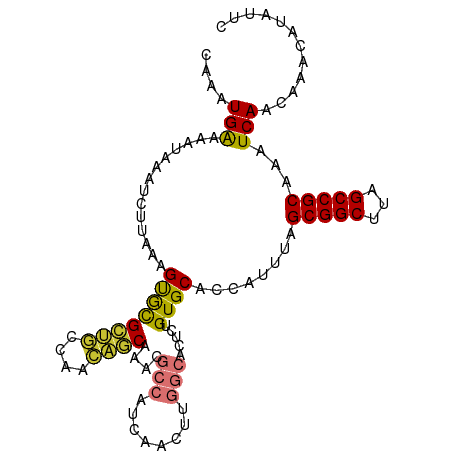
>dm3.chr3R 24208034 102 - 27905053 AAUGAAAAUAAAUCUUAAAGUGCG-CUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- ..(((..............(((((-(((....))))....((((......)))).......))))......(((((...)))))...))).............- ( -27.20, z-score = -3.19, R) >droSim1.chr3R 23894919 102 - 27517382 AAUGAAAAUAAAUCUUAAAGUGCG-CUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- ..(((..............(((((-(((....))))....((((......)))).......))))......(((((...)))))...))).............- ( -27.20, z-score = -3.19, R) >droSec1.super_22 811489 102 - 1066717 AAUGAAAAUAAAUCUUAAAGUGCG-CUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- ..(((..............(((((-(((....))))....((((......)))).......))))......(((((...)))))...))).............- ( -27.20, z-score = -3.19, R) >droYak2.chr3R 6538944 102 + 28832112 AAUGAAAAUAAAUCUUAAAGUGCG-CUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- ..(((..............(((((-(((....))))....((((......)))).......))))......(((((...)))))...))).............- ( -27.20, z-score = -3.19, R) >droEre2.scaffold_4820 6635681 102 + 10470090 AAUGAAAAUAAAUCUUAAAGUGCG-CUGCCAACAGCAAACGCCAUCAACUUGGCACUCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- ..(((..............(((((-(((....))))....((((......)))).......))))......(((((...)))))...))).............- ( -27.20, z-score = -3.36, R) >droAna3.scaffold_13340 17008660 99 - 23697760 AAUGAAAAUAAAUCUUAAAGUAUGCUCACCCACAAUGGCCAUCAACU-----CGGCGCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUACGC ...................(((((......((((...(((.......-----.)))..)))).........(((((...))))).............))))).. ( -18.60, z-score = -0.39, R) >dp4.chr2 20467910 89 + 30794189 --UGGAAAUAAAUCUUAAAGCACG-UC----AAAGCUGACGACGACA-------AUGCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- --..(((............((.((-((----(....)))))(((...-------....)))))........(((((...)))))...............))).- ( -18.50, z-score = -1.09, R) >droPer1.super_0 10155413 89 - 11822988 --UGGAAAUAAAUCUUAAAGCACG-UC----AAAGCUGACGACGACA-------AUGCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG- --..(((............((.((-((----(....)))))(((...-------....)))))........(((((...)))))...............))).- ( -18.50, z-score = -1.09, R) >consensus AAUGAAAAUAAAUCUUAAAGUGCG_CUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUCG_ ..(((..............(((((..(((((...................)))))...)))))........(((((...)))))...))).............. (-15.48 = -15.37 + -0.11)
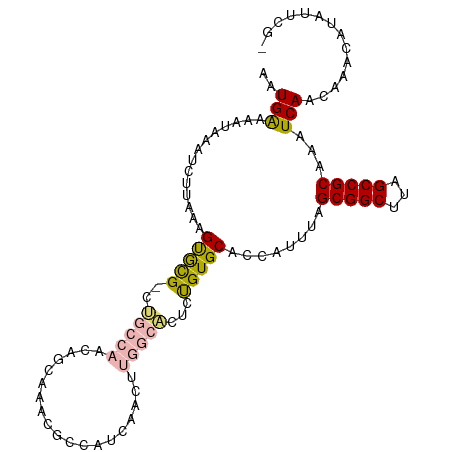
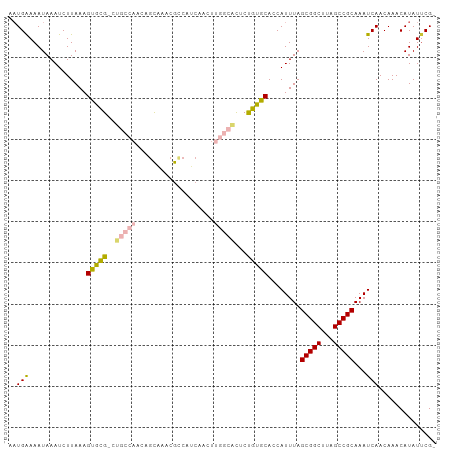
| Location | 24,208,035 – 24,208,138 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.54 |
| Shannon entropy | 0.31613 |
| G+C content | 0.42957 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -22.03 |
| Energy contribution | -21.99 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24208035 103 + 27905053 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACAGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((.(((..(.((((..((((((...)))(((...(((..(.....)..))).((..(((((....)))))..))))))))..)))).)..))).)))..... ( -32.70, z-score = -1.78, R) >droSim1.chr3R 23894920 103 + 27517382 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACAGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((.(((..(.((((..((((((...)))(((...(((..(.....)..))).((..(((((....)))))..))))))))..)))).)..))).)))..... ( -32.70, z-score = -1.78, R) >droSec1.super_22 811490 103 + 1066717 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACAGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((.(((..(.((((..((((((...)))(((...(((..(.....)..))).((..(((((....)))))..))))))))..)))).)..))).)))..... ( -32.70, z-score = -1.78, R) >droYak2.chr3R 6538945 103 - 28832112 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACAGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((.(((..(.((((..((((((...)))(((...(((..(.....)..))).((..(((((....)))))..))))))))..)))).)..))).)))..... ( -32.70, z-score = -1.78, R) >droEre2.scaffold_4820 6635682 103 - 10470090 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACGGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((.(((..(.((((..((((((...)))(((...(((..(.....)..))).((..(((((....)))))..))))))))..)))).)..))).)))..... ( -32.70, z-score = -1.70, R) >droAna3.scaffold_13340 17008662 99 + 23697760 GUAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCAC--AGCGCCGAGUUGAUGGCCAUUGUGGGUG--AGCAUACUUUAAGAUUUAUUUUCAUUUG ...((((((..((.((..(((((...)))))..((((((.(((--(((.....)))).)))))))))).))..--))))))...................... ( -26.30, z-score = -0.54, R) >dp4.chr2 20467911 90 - 30794189 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCAC--GGC---------AUUGUCGUCGUCAGCUUUGACGUGCUUUAAGAUUUAUUUCCAUG-- ...((((((.((((((..(((((...(((((........))..--)))---------...))))).))))))...))))))....................-- ( -22.20, z-score = -0.30, R) >droPer1.super_0 10155414 90 + 11822988 GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCAC--GGC---------AUUGUCGUCGUCAGCUUUGACGUGCUUUAAGAUUUAUUUCCAUG-- ...((((((.((((((..(((((...(((((........))..--)))---------...))))).))))))...))))))....................-- ( -22.20, z-score = -0.30, R) >consensus GAAUAUGUUUGUUGAUUUGCGGCUAAGCCGCUAAAUGGUGCACAGAGUGCCAAGUUGAUGGCGUUUGCUGUUGGCAGCGCACUUUAAGAUUUAUUUUCAUUUG (((((.((((....(((((((((...))))).))))(((((.......((((......))))....((((....)))))))))...)))).)))))....... (-22.03 = -21.99 + -0.04)
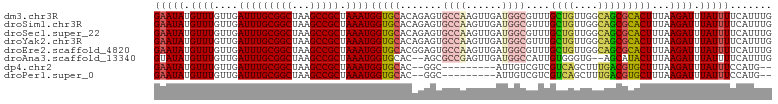
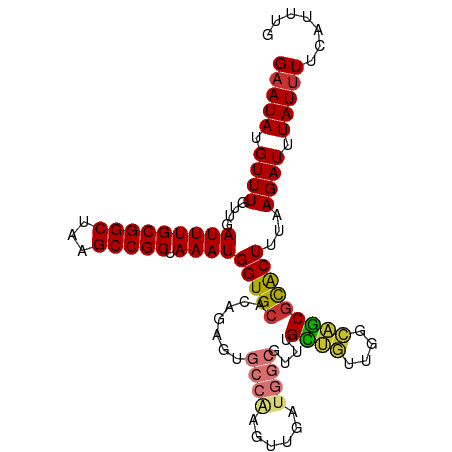
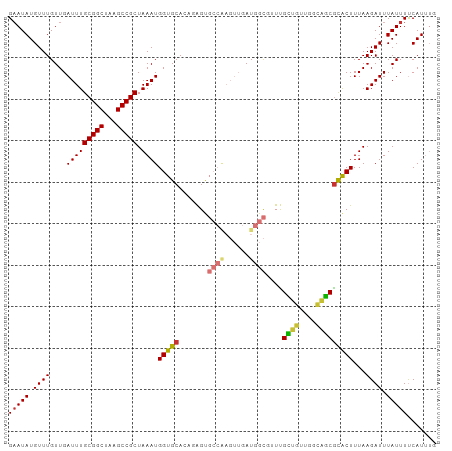
| Location | 24,208,035 – 24,208,138 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.54 |
| Shannon entropy | 0.31613 |
| G+C content | 0.42957 |
| Mean single sequence MFE | -23.89 |
| Consensus MFE | -20.10 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24208035 103 - 27905053 CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....((((......)))).......))))......(((((...)))))...)))............ ( -27.20, z-score = -3.21, R) >droSim1.chr3R 23894920 103 - 27517382 CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....((((......)))).......))))......(((((...)))))...)))............ ( -27.20, z-score = -3.21, R) >droSec1.super_22 811490 103 - 1066717 CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....((((......)))).......))))......(((((...)))))...)))............ ( -27.20, z-score = -3.21, R) >droYak2.chr3R 6538945 103 + 28832112 CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....((((......)))).......))))......(((((...)))))...)))............ ( -27.20, z-score = -3.21, R) >droEre2.scaffold_4820 6635682 103 + 10470090 CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCCGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....((((......)))).......))))......(((((...)))))...)))............ ( -27.20, z-score = -3.43, R) >droAna3.scaffold_13340 17008662 99 - 23697760 CAAAUGAAAAUAAAUCUUAAAGUAUGCU--CACCCACAAUGGCCAUCAACUCGGCGCU--GUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUAC .....................(((((..--....((((...(((........)))..)--))).........(((((...)))))...........))))).. ( -18.10, z-score = -0.76, R) >dp4.chr2 20467911 90 + 30794189 --CAUGGAAAUAAAUCUUAAAGCACGUCAAAGCUGACGACGACAAU---------GCC--GUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC --.((((.................(((((....)))))(((.....---------..)--))...))))...(((((...))))).................. ( -18.50, z-score = -1.29, R) >droPer1.super_0 10155414 90 - 11822988 --CAUGGAAAUAAAUCUUAAAGCACGUCAAAGCUGACGACGACAAU---------GCC--GUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC --.((((.................(((((....)))))(((.....---------..)--))...))))...(((((...))))).................. ( -18.50, z-score = -1.29, R) >consensus CAAAUGAAAAUAAAUCUUAAAGUGCGCUGCCAACAGCAAACGCCAUCAACUUGGCACUCUGUGCACCAUUUAGCGGCUUAGCCGCAAAUCAACAAACAUAUUC ....(((..............((((((((....))))....(((........))).....))))........(((((...)))))...)))............ (-20.10 = -19.57 + -0.53)
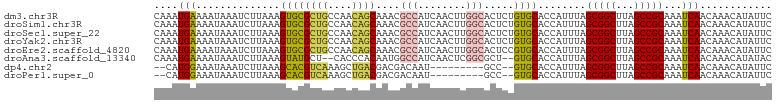
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:49 2011