
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,203,144 – 24,203,235 |
| Length | 91 |
| Max. P | 0.754381 |

| Location | 24,203,144 – 24,203,235 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.15 |
| Shannon entropy | 0.23937 |
| G+C content | 0.43744 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
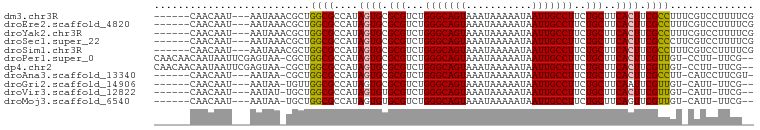
>dm3.chr3R 24203144 91 + 27905053 ------CAACAAU---AAUAAACGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCUUUCGUCCUUUUCG ------.......---.....(((..((((....((((.(((...(((((((...........)))))))..)))..)))).))))...)))........ ( -21.50, z-score = -1.94, R) >droEre2.scaffold_4820 6630690 91 - 10470090 ------CAACAAU---AAUAAACGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCUUUCGUCCUUUUCG ------.......---.....(((..((((....((((.(((...(((((((...........)))))))..)))..)))).))))...)))........ ( -21.50, z-score = -1.94, R) >droYak2.chr3R 6533884 91 - 28832112 ------CAACAAU---AAUAAACGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCUUUCGUCCUUUUCG ------.......---.....(((..((((....((((.(((...(((((((...........)))))))..)))..)))).))))...)))........ ( -21.50, z-score = -1.94, R) >droSec1.super_22 806652 91 + 1066717 ------CAACAAU---AAUAAACGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCCUUCGUCCUUUUCG ------.......---.....(((..((.((...((((.(((...(((((((...........)))))))..)))..))))..))))..)))........ ( -20.90, z-score = -1.75, R) >droSim1.chr3R 23890032 91 + 27517382 ------CAACAAU---AAUAAACGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCUUUCGUCCUUUUCG ------.......---.....(((..((((....((((.(((...(((((((...........)))))))..)))..)))).))))...)))........ ( -21.50, z-score = -1.94, R) >droPer1.super_0 10150808 95 + 11822988 CAACAACAAUAAUUCGAGUAA-CGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGUUGU-CCUU-UUCG-- ...............(..(((-((..(((((....))))(((...(((((((...........)))))))..)))....)..))))).-.)..-....-- ( -20.00, z-score = -0.58, R) >dp4.chr2 20463251 95 - 30794189 CAACAACAAUAAUUCGAGUAA-CGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGUUGU-CCUU-UUCG-- ...............(..(((-((..(((((....))))(((...(((((((...........)))))))..)))....)..))))).-.)..-....-- ( -20.00, z-score = -0.58, R) >droAna3.scaffold_13340 17003987 88 + 23697760 ------CAACAAU---AAUAA-CGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCUU-CAUCCUUCGU- ------.......---.....-.(..((((....((((.(((...(((((((...........)))))))..)))..)))).))))..-).........- ( -20.30, z-score = -1.50, R) >droGri2.scaffold_14906 1233429 86 + 14172833 ------CAACAAU---AAUAA-UGUUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCAAUUCGUUGU-CAUU-UUCG-- ------((((...---.....-.....((((....))))(((...(((((((...........)))))))..)))........)))).-....-....-- ( -17.70, z-score = -0.98, R) >droVir3.scaffold_12822 3622498 86 + 4096053 ------CAACAAU---AAUAU-UGCUGGCGCCAUAGUGUGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGUUGU-CAUU-UUCG-- ------.......---.....-...(((((.(..((((.(((...(((((((...........)))))))..)))..))))..).)))-))..-....-- ( -18.90, z-score = -1.32, R) >droMoj3.scaffold_6540 16937717 86 + 34148556 ------CAACAAU---AAUAA-UGCUGGCGCCAUAGUGUGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCAGUUCGUUGU-CAUU-UUCG-- ------.....((---(((..-.((((((((....))))(((...(((((((...........)))))))..)))..))))..)))))-....-....-- ( -17.30, z-score = -0.51, R) >consensus ______CAACAAU___AAUAA_CGCUGGCGCCAUAGUGCGCGUCUGGGCAGUAAAUAAAAAUAAUUGCCUUCUGCUUCACUUCGCCGU_CGUCCUUCG__ ..........................((((....((((.(((...(((((((...........)))))))..)))..)))).)))).............. (-17.43 = -17.12 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:45 2011