
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,176,168 – 24,176,311 |
| Length | 143 |
| Max. P | 0.888714 |

| Location | 24,176,168 – 24,176,286 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.37 |
| Shannon entropy | 0.26351 |
| G+C content | 0.39726 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -18.71 |
| Energy contribution | -18.49 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24176168 118 + 27905053 --CGCCGGCGAGCGGCAAAUGCAAAAAGUUUCCACUCAAGAAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAACCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --.((((.....))))(((((((...(((....)))....(((((((((((......(((((.......))))).....((....)))))))))))))..)))))))............. ( -31.10, z-score = -1.94, R) >droSim1.chr3R 23858466 118 + 27517382 --CGCCGGCGAGCGGCAAAUGCAAAAAGUUUCCACUCAAGAAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --.((((.....))))(((((((...(((....)))....(((((((((((......(((((.......)))))(((....)))...)))))))))))..)))))))............. ( -29.90, z-score = -1.26, R) >droSec1.super_22 779900 118 + 1066717 --CGCCGGCGAGUGGCAAAUGCAAAAAGUUUCCACUCAAGAAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --.......((((((.((((.......))))))))))..((((((((((((......(((((.......)))))(((....)))...)))))))((((((......))))))..))))). ( -31.60, z-score = -1.87, R) >droEre2.scaffold_4820 6599835 112 - 10470090 --CGCCAGCGAG---CAAAUGCAAAAAGUUUCCACUCAA---GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --.((......)---)(((((((...(((....))).((---(((((((((......(((((.......)))))(((....)))...)))))))))))..)))))))............. ( -23.90, z-score = -0.93, R) >droYak2.chr3R 6505884 112 - 28832112 --CACCAGCGAG---CAAAUGCAAAAAGUUUCCACUCAA---GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --..........---.(((((((...(((....))).((---(((((((((......(((((.......)))))(((....)))...)))))))))))..)))))))............. ( -22.90, z-score = -1.00, R) >droAna3.scaffold_13340 16977853 103 + 23697760 --------------GCAAAUGCAAAAAGUUUCCACUCAA---GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --------------..(((((((...(((....))).((---(((((((((......(((((.......)))))(((....)))...)))))))))))..)))))))............. ( -22.90, z-score = -1.97, R) >dp4.chr2 20433662 107 - 30794189 ---------GAG-AGCAAAUGCAAAAAGUUUCCACUCAA---GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCCUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC ---------(((-((.(((((((...(((....))).((---(((((((((......(((((.......)))))(((....)))...)))))))))))..))))))).......))))). ( -24.20, z-score = -2.02, R) >droPer1.super_0 10121206 107 + 11822988 ---------GAG-AGCAAAUGCAAAAAGUUUCCACUCAA---GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCCUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC ---------(((-((.(((((((...(((....))).((---(((((((((......(((((.......)))))(((....)))...)))))))))))..))))))).......))))). ( -24.20, z-score = -2.02, R) >droWil1.scaffold_181089 8827165 116 + 12369635 UUCUUUCUACCCAGGAAAAUGUAAAAAGUUUCCACUCAA----GAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC .............(((((.(((..(((((...((.(.((----((((((((......((((((.....))))))(((....)))...)))))))))).).)).)))))...))).))))) ( -26.80, z-score = -3.79, R) >droVir3.scaffold_12822 3586236 102 + 4096053 --------------GCAAAUGUAAAAAGUUUCCACUCUA----GAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --------------.....(((..(((((...((....(----((((((((......((((((.....))))))(((....)))...)))))))))....)).)))))...)))...... ( -20.60, z-score = -2.40, R) >droMoj3.scaffold_6540 16901647 94 + 34148556 --------------GCAAAUGUAAAAAGUUUGCACUCUA----GAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUAGCAGCCGUGAACUUUUUAAUGUAUUUUAAAC-------- --------------((((((.......))))))((...(----((((((((......((((((.....))))))(((....)))...))))))))).....)).........-------- ( -23.00, z-score = -3.12, R) >droGri2.scaffold_14906 1202419 102 + 14172833 --------------GCAAAUGUAAAAAGUUUCCACUCUC----GAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCACGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC --------------(((....(((((((((..(((....----(((((.....)))))(((((.....)))))))).(((((...)))))))))))))).)))................. ( -21.00, z-score = -2.61, R) >consensus _________GAG__GCAAAUGCAAAAAGUUUCCACUCAA___GGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCC ................((((((((((((((..(((((......))))..........(((((.......)))))(((....)))...)..)))))))...)))))))............. (-18.71 = -18.49 + -0.22)

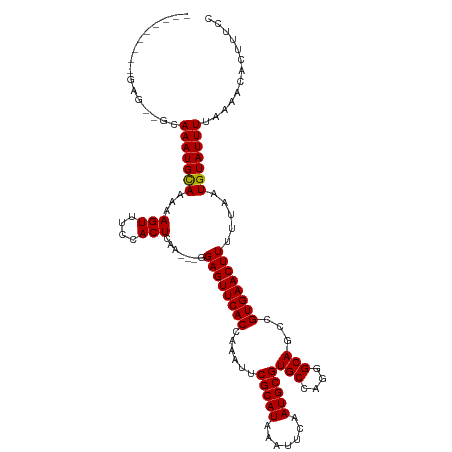

| Location | 24,176,206 – 24,176,311 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Shannon entropy | 0.14313 |
| G+C content | 0.35882 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24176206 105 + 27905053 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAACCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU------- ..(((((((((......(((((.......))))).....((....)))))))).((((((......)))))).....)))............((((.....))))------- ( -23.10, z-score = -2.47, R) >droSim1.chr3R 23858504 105 + 27517382 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU------- ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))............((((.....))))------- ( -21.90, z-score = -1.64, R) >droSec1.super_22 779938 105 + 1066717 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU------- ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))............((((.....))))------- ( -21.90, z-score = -1.64, R) >droEre2.scaffold_4820 6599867 105 - 10470090 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU------- ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))............((((.....))))------- ( -21.90, z-score = -1.64, R) >droYak2.chr3R 6505916 105 - 28832112 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU------- ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))............((((.....))))------- ( -21.90, z-score = -1.64, R) >droAna3.scaffold_13340 16977876 105 + 23697760 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACGGCUU------- ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))............((((.....))))------- ( -21.50, z-score = -1.36, R) >droPer1.super_0 10121233 112 + 11822988 AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCCUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUUGCCUCUU ..(((((((((......(((((.......)))))(((....)))...)))))).((((((......)))))).....)))...........(((((.....)))))...... ( -23.40, z-score = -1.98, R) >droWil1.scaffold_181089 8827202 104 + 12369635 -AAGAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCGU------- -((((((((((......((((((.....))))))(((....)))...)))))))))).....................................((.....))..------- ( -21.50, z-score = -1.95, R) >droVir3.scaffold_12822 3586259 104 + 4096053 -UAGAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGUAAACUGCUC------- -.(((((((((......((((((.....))))))(((....)))...))))))))).................................................------- ( -19.30, z-score = -1.78, R) >droMoj3.scaffold_6540 16901670 96 + 34148556 -UAGAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCAUAGCAGCCGUGAACUUUUUAAUGUAUUUUAAA--------CACUAAACAUUUUAAGUAAACUGCUG------- -.(((((((((......((((((.....))))))(((....)))...)))))))))...............--------..........................------- ( -17.40, z-score = -1.47, R) >droGri2.scaffold_14906 1202442 104 + 14172833 -UCGAGUUCACCAAAUUCGCAUAAAUUCUAUGCGUGCCACGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGUAAACUGCUC------- -..((((((((......((((((.....))))))(((....)))...))))))))(((((......)))))..................................------- ( -19.00, z-score = -1.88, R) >consensus AAGGAGUUCACCAAAUUCGCAUAAAUUCAAUGCGUGCCAGGGCAGCCGUGAACUUUUUAAUGUAUUUUAAAACACUUUCCACUAAACAUUUUAAGCAAACUGCUU_______ ..(((((((((......(((((.......)))))(((....)))...))))))))).....................................(((.....)))........ (-18.43 = -18.25 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:41 2011