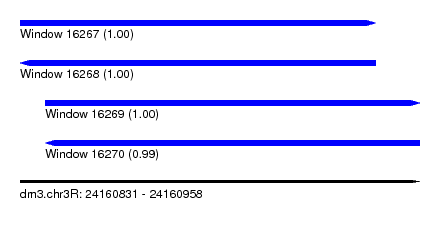
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,160,831 – 24,160,958 |
| Length | 127 |
| Max. P | 0.999642 |

| Location | 24,160,831 – 24,160,944 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.58 |
| Shannon entropy | 0.48929 |
| G+C content | 0.38837 |
| Mean single sequence MFE | -31.89 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.97 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24160831 113 + 27905053 ------CUAGGAUUUUCCAACAAGAUUGCGAGUUGGGCGAACGCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC-UUCUGCAAUAAUAUUUACAUCUUUACUUAA ------...((.....))......(((((.((..((((((((..(((((....((((((.......))))))..)))))..))))))))-..)))))))..................... ( -35.50, z-score = -3.76, R) >droSim1.chr3R 23843214 113 + 27517382 ------CUAGGAUUUUCAAAAAAGAUUGCGAGUUUGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC-UCCUGCAAUAAUAUUUACAUCUUUACUUAA ------..(((((...........((((((.(...((((((((.(((((....((((((.......))))))..))))).)))))))).-.).)))))).........)))))....... ( -31.85, z-score = -3.23, R) >droSec1.super_22 764625 109 + 1066717 ------CUAGGAUUUUCAAAU----UUGCGAGUUUGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGUUUAAUCUCAAGUUCGCCC-UCCUGCAAUAAUAUUUACAUCUUUACUUAA ------..(((((........----(((((.(...((((((((.(((((...(((((........)))))....))))).)))))))).-.).)))))..........)))))....... ( -28.27, z-score = -2.71, R) >droYak2.chr3R 6490021 110 - 28832112 ------CUAGGAUUUUCCAACUAGAUUGCGAGAUGGGCGAACUCGAGGUGUUUGUUGCUCAAUUACAGUGAUUAAUCUCAAGUUCGCCCCUCCUGCAUAAAUAUUUA----UUUACUUAA ------((((..........))))..((((.((.(((((((((.(((((....((..((.......))..))..))))).))))))))).)).))))..........----......... ( -35.20, z-score = -4.38, R) >droEre2.scaffold_4820 6583843 110 - 10470090 ------CCAGGAUUUUCCAACUAGAUUGCGAGAUGGGCGAACUCGAGGUGUUUGUUGCCCAAUUACUGCGAUUAAUCUCAAGUUCGCCCCUCCUGCAAUACUAUUUA----UUUACUUAA ------...((.....))...(((((((((.((.(((((((((.(((((....(((((.........)))))..))))).))))))))).)).)))))).)))....----......... ( -38.60, z-score = -5.52, R) >droMoj3.scaffold_6498 2595538 115 - 3408170 CUAUUAUAUGAAUGUACAUGCAAUAGCAUAGCCUUAGAGAUGGCAUGUUGCCCAAAGCUUAACCACGCUUACCUAUCU--AAUUCGAAC---UUGCAUUCACAAAUACAUAUAUAAUUUA ....((((((..(((..((((((((((((.(((........)))))))))....((((........))))........--.........---))))))..)))....))))))....... ( -21.90, z-score = -1.49, R) >consensus ______CUAGGAUUUUCAAACAAGAUUGCGAGUUGGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC_UCCUGCAAUAAUAUUUACAUCUUUACUUAA ........................((((((.....((((((((.(((((....((((((.......))))))..))))).)))))))).....))))))..................... (-18.74 = -20.97 + 2.23)


| Location | 24,160,831 – 24,160,944 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.58 |
| Shannon entropy | 0.48929 |
| G+C content | 0.38837 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -14.80 |
| Energy contribution | -16.86 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.997405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24160831 113 - 27905053 UUAAGUAAAGAUGUAAAUAUUAUUGCAGAA-GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGCGUUCGCCCAACUCGCAAUCUUGUUGGAAAAUCCUAG------ .........(((...((((..(((((((..-((((((((.((((.......((((....)).)).........)))).))))))))..)).)))))..)))).....)))....------ ( -29.79, z-score = -2.72, R) >droSim1.chr3R 23843214 113 - 27517382 UUAAGUAAAGAUGUAAAUAUUAUUGCAGGA-GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAAUCUUUUUUGAAAAUCCUAG------ .........(((.((((.(..(((((((..-.((((((((((((.......((((....)).)).........))))))))))))...)).)))))..).))))...)))....------ ( -30.29, z-score = -3.24, R) >droSec1.super_22 764625 109 - 1066717 UUAAGUAAAGAUGUAAAUAUUAUUGCAGGA-GGGCGAACUUGAGAUUAAACGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAA----AUUUGAAAAUCCUAG------ .........(((.(((((....((((((..-.((((((((((((.......((((....)).)).........))))))))))))...)).))))----)))))...)))....------ ( -28.59, z-score = -3.40, R) >droYak2.chr3R 6490021 110 + 28832112 UUAAGUAAA----UAAAUAUUUAUGCAGGAGGGGCGAACUUGAGAUUAAUCACUGUAAUUGAGCAACAAACACCUCGAGUUCGCCCAUCUCGCAAUCUAGUUGGAAAAUCCUAG------ .((((((..----....))))))(((..((.(((((((((((((.........(((......)))........))))))))))))).))..)))........((.....))...------ ( -32.83, z-score = -4.06, R) >droEre2.scaffold_4820 6583843 110 + 10470090 UUAAGUAAA----UAAAUAGUAUUGCAGGAGGGGCGAACUUGAGAUUAAUCGCAGUAAUUGGGCAACAAACACCUCGAGUUCGCCCAUCUCGCAAUCUAGUUGGAAAAUCCUGG------ .........----....(((.(((((..((.(((((((((((((........(((...)))(....)......))))))))))))).))..))))))))...((.....))...------ ( -36.50, z-score = -4.08, R) >droMoj3.scaffold_6498 2595538 115 + 3408170 UAAAUUAUAUAUGUAUUUGUGAAUGCAA---GUUCGAAUU--AGAUAGGUAAGCGUGGUUAAGCUUUGGGCAACAUGCCAUCUCUAAGGCUAUGCUAUUGCAUGUACAUUCAUAUAAUAG .......((((((....((((.((((((---..((.....--.))......(((((((((........(((.....)))........))))))))).)))))).))))..)))))).... ( -29.09, z-score = -1.38, R) >consensus UUAAGUAAAGAUGUAAAUAUUAUUGCAGGA_GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAAUCUUGUUGGAAAAUCCUAG______ .......................(((.((...((((((((((((..........((......)).........))))))))))))...)).))).......................... (-14.80 = -16.86 + 2.06)

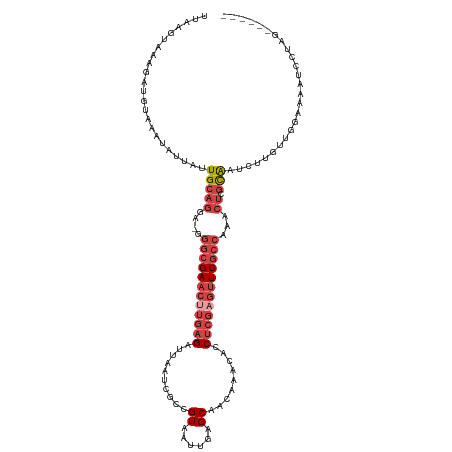
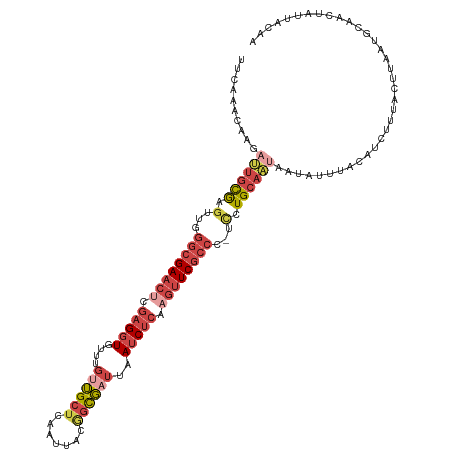
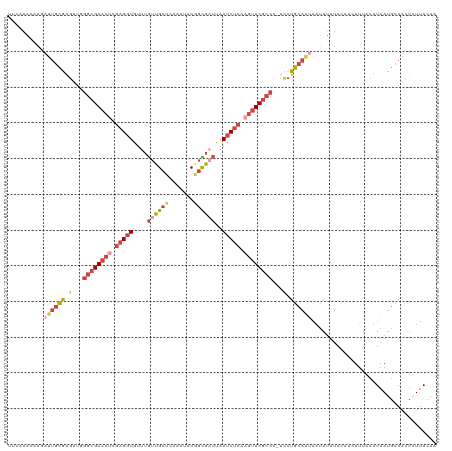
| Location | 24,160,839 – 24,160,958 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.47040 |
| G+C content | 0.38707 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -18.74 |
| Energy contribution | -20.97 |
| Covariance contribution | 2.23 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24160839 119 + 27905053 UUCCAACAAGAUUGCGAGUUGGGCGAACGCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC-UUCUGCAAUAAUAUUUACAUCUUUACUUAAUGCAACUAUUACAG ..........(((((.((..((((((((..(((((....((((((.......))))))..)))))..))))))))-..)))))))................................... ( -33.80, z-score = -3.30, R) >droSim1.chr3R 23843222 119 + 27517382 UUCAAAAAAGAUUGCGAGUUUGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC-UCCUGCAAUAAUAUUUACAUCUUUACUUAAUGCAACUAUUACAC ..........((((((.(...((((((((.(((((....((((((.......))))))..))))).)))))))).-.).))))))................................... ( -31.00, z-score = -3.37, R) >droSec1.super_22 764633 115 + 1066717 UUCAAAU----UUGCGAGUUUGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGUUUAAUCUCAAGUUCGCCC-UCCUGCAAUAAUAUUUACAUCUUUACUUAAUGCAACUAUUACAC .......----(((((.(...((((((((.(((((...(((((........)))))....))))).)))))))).-.).))))).................................... ( -27.40, z-score = -2.67, R) >droYak2.chr3R 6490029 116 - 28832112 UUCCAACUAGAUUGCGAGAUGGGCGAACUCGAGGUGUUUGUUGCUCAAUUACAGUGAUUAAUCUCAAGUUCGCCCCUCCUGCAUAAAUAUUUA----UUUACUUAAUGCAACUAUUACAA .......(((.(((((((..(((((((((.(((((....((..((.......))..))..))))).)))))))))))).....(((((....)----))))......)))))))...... ( -33.00, z-score = -3.99, R) >droEre2.scaffold_4820 6583851 116 - 10470090 UUCCAACUAGAUUGCGAGAUGGGCGAACUCGAGGUGUUUGUUGCCCAAUUACUGCGAUUAAUCUCAAGUUCGCCCCUCCUGCAAUACUAUUUA----UUUACUUAAUGCAACUGUUACAA .......(((((((((.((.(((((((((.(((((....(((((.........)))))..))))).))))))))).)).)))))).)))....----....................... ( -36.90, z-score = -5.07, R) >droMoj3.scaffold_6498 2595552 115 - 3408170 UACAUGCAAUAGCAUAGCCUUAGAGAUGGCAUGUUGCCCAAAGCUUAACCACGCUUACCUAUCU--AAUUCGAAC---UUGCAUUCACAAAUACAUAUAUAAUUUACACAGCCAUUAAAG ...((((((((((((.(((........)))))))))....((((........))))........--.........---)))))).................................... ( -16.80, z-score = -0.61, R) >consensus UUCAAACAAGAUUGCGAGUUGGGCGAACUCGAGGUGUUUGUUGCUCAAUUACGGCGAUUAAUCUCAAGUUCGCCC_UCCUGCAAUAAUAUUUACAUCUUUACUUAAUGCAACUAUUACAA ..........((((((.....((((((((.(((((....((((((.......))))))..))))).)))))))).....))))))................................... (-18.74 = -20.97 + 2.23)

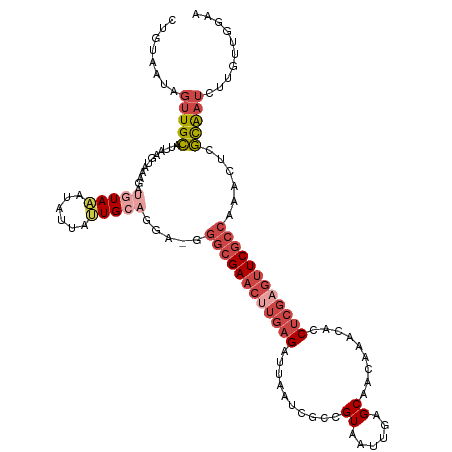
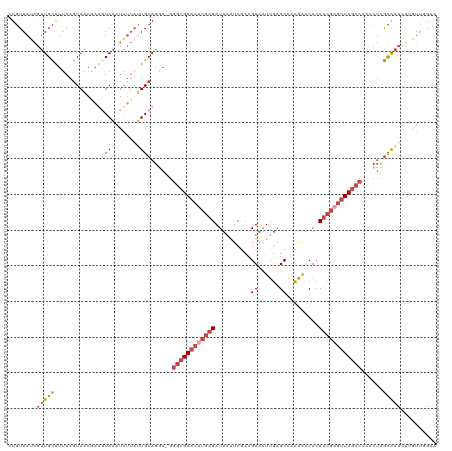
| Location | 24,160,839 – 24,160,958 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Shannon entropy | 0.47040 |
| G+C content | 0.38707 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -16.02 |
| Energy contribution | -18.08 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24160839 119 - 27905053 CUGUAAUAGUUGCAUUAAGUAAAGAUGUAAAUAUUAUUGCAGAA-GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGCGUUCGCCCAACUCGCAAUCUUGUUGGAA ((((((((((((((((.......))))))...))))))))))..-((((((((.((((.......((((....)).)).........)))).))))))))..((.(((....))).)).. ( -34.19, z-score = -2.92, R) >droSim1.chr3R 23843222 119 - 27517382 GUGUAAUAGUUGCAUUAAGUAAAGAUGUAAAUAUUAUUGCAGGA-GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAAUCUUUUUUGAA ..((((((.(((((((.......))))))).))))))(((((..-.((((((((((((.......((((....)).)).........))))))))))))...)).)))............ ( -33.89, z-score = -3.26, R) >droSec1.super_22 764633 115 - 1066717 GUGUAAUAGUUGCAUUAAGUAAAGAUGUAAAUAUUAUUGCAGGA-GGGCGAACUUGAGAUUAAACGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAA----AUUUGAA ...(((((.(((((((.......))))))).)))))((((((..-.((((((((((((.......((((....)).)).........))))))))))))...)).))))----....... ( -33.99, z-score = -3.89, R) >droYak2.chr3R 6490029 116 + 28832112 UUGUAAUAGUUGCAUUAAGUAAA----UAAAUAUUUAUGCAGGAGGGGCGAACUUGAGAUUAAUCACUGUAAUUGAGCAACAAACACCUCGAGUUCGCCCAUCUCGCAAUCUAGUUGGAA ((((.....((((((.(((((..----....)))))))))))((((((((((((((((.........(((......)))........)))))))))))))..)))))))........... ( -33.93, z-score = -3.47, R) >droEre2.scaffold_4820 6583851 116 + 10470090 UUGUAACAGUUGCAUUAAGUAAA----UAAAUAGUAUUGCAGGAGGGGCGAACUUGAGAUUAAUCGCAGUAAUUGGGCAACAAACACCUCGAGUUCGCCCAUCUCGCAAUCUAGUUGGAA .(.((((..((((.....)))).----....(((.(((((..((.(((((((((((((........(((...)))(....)......))))))))))))).))..)))))))))))).). ( -38.60, z-score = -4.26, R) >droMoj3.scaffold_6498 2595552 115 + 3408170 CUUUAAUGGCUGUGUAAAUUAUAUAUGUAUUUGUGAAUGCAA---GUUCGAAUU--AGAUAGGUAAGCGUGGUUAAGCUUUGGGCAACAUGCCAUCUCUAAGGCUAUGCUAUUGCAUGUA ........((.((((((......(((.(((((((((((....---)))))...)--))))).)))(((((((((........(((.....)))........))))))))).)))))))). ( -25.89, z-score = 0.49, R) >consensus CUGUAAUAGUUGCAUUAAGUAAAGAUGUAAAUAUUAUUGCAGGA_GGGCGAACUUGAGAUUAAUCGCCGUAAUUGAGCAACAAACACCUCGAGUUCGCCAAACUCGCAAUCUUGUUGGAA ........(((((............(((((......))))).....((((((((((((..........((......)).........))))))))))))......))))).......... (-16.02 = -18.08 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:37 2011