
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,138,526 – 24,138,623 |
| Length | 97 |
| Max. P | 0.708547 |

| Location | 24,138,526 – 24,138,623 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.19 |
| Shannon entropy | 0.47506 |
| G+C content | 0.51845 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -15.74 |
| Energy contribution | -15.34 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.708547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
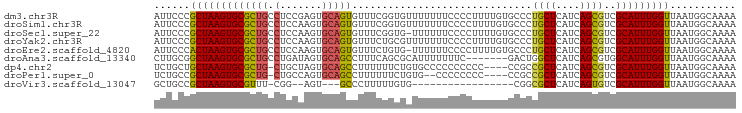
>dm3.chr3R 24138526 97 - 27905053 AUUCCCGCUAAGUGCGCUGCCUCCGAGUGCAGUGUUUCGGUGUUUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA .....(((.....))).((((.((((((((........((..(..............)..))..(((....)))...)))))))).....))))... ( -23.94, z-score = -0.75, R) >droSim1.chr3R 23823311 97 - 27517382 AUUCCCGCUAAGUGCGCUGCCUCCAAGUGCAGUGUUUCGGUGUUUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA .....(((.....))).((((.((((((((........((..(..............)..))..(((....)))...)))))))).....))))... ( -24.24, z-score = -1.13, R) >droSec1.super_22 742243 96 - 1066717 AUUCCCGCUAAGUGCGCUGCCUCCAAGUGCAGUGUUUCGGUG-UUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA .....(((.....))).((((.((((((((........((..-(.............)..))..(((....)))...)))))))).....))))... ( -24.32, z-score = -1.09, R) >droYak2.chr3R 6466918 97 + 28832112 AUUCCCGCUAAGUGCGCUGCCUCCAAGUGCAGUGUUUCUGCGUUUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA .....(((.....))).((((.((((((((((.....))))..................((..((((....))))..)))))))).....))))... ( -23.50, z-score = -1.09, R) >droEre2.scaffold_4820 6560493 96 + 10470090 AUUCCCACUAAGUGCGCUGCCUCCAAGUGCAGUGUUUCUGUG-UUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA ......((((((((((.(((....(((..(((.....)))..-)))..........((.((...)).))...))).))))))))))........... ( -21.50, z-score = -1.02, R) >droAna3.scaffold_13340 22263651 90 - 23697760 CUUGCGGCUAAGUGCGCUGCCUGAUAGUGCAGCCUUUCAGCGCAUUUUUUUC-------GACUGGCUCAUCAGCGUGGCAUUUGGUUAAUGGCAAAA .((((((((((((((((((((.....).)))))....(((((.(.....).)-------).)))(((....)))...))))))))))....)))).. ( -25.30, z-score = 0.99, R) >dp4.chr2 22848055 92 + 30794189 UCUGCUGCUAAGUGCGCUG-CUGCUAGUGCAGCCUUUUUUCUGUGCCCCCCCCCC----CCGCCGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA ..((((((((((((((..(-((((....)))))......................----....((((....)))).))))))))))....))))... ( -27.20, z-score = -1.46, R) >droPer1.super_0 10042827 90 + 11822988 UCUGCCGCUAAGUGCGCUG-CUGCCAGUGCAGCCUUUUUUCUGUG--CCCCCCCC----CCGCCGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA ..(((((((((((((((((-(.......)))))............--........----..(.((((....)))).))))))))))....))))... ( -28.70, z-score = -1.88, R) >droVir3.scaffold_13047 3807900 74 + 19223366 GCUGCCGCUAAGUGCGUUU-CGG--AGU---GCCCUUUUUGUG-----------------CGGCGCUCAUCAGUGUCGCAUUUGGUUAAUGGCAAAA ..((((((((((((((...-(.(--(((---((((.......)-----------------.)))))))....)...))))))))))....))))... ( -30.50, z-score = -2.95, R) >consensus AUUCCCGCUAAGUGCGCUGCCUCCAAGUGCAGUGUUUCUGCGGUUUUUUCCCCUUUUGUGCCCUGCUCAUCAGCGUCGCAUUUGGUUAAUGGCAAAA ......(((((((((((((..........))))..............................((((....))))..)))))))))........... (-15.74 = -15.34 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:32 2011