| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,128,566 – 24,128,664 |
| Length | 98 |
| Max. P | 0.899879 |

| Location | 24,128,566 – 24,128,664 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.50 |
| Shannon entropy | 0.41138 |
| G+C content | 0.56553 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -27.10 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
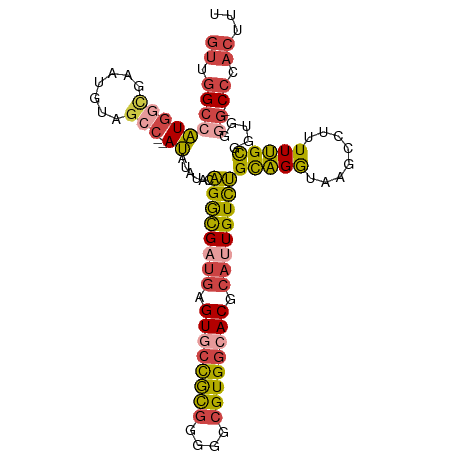
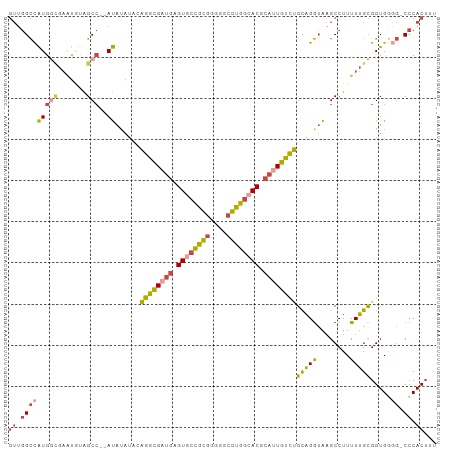
>dm3.chr3R 24128566 98 - 27905053 GUCGGCCAUGGCGAAUAUAGCC--AUAUAUAGAGGCGUUGAGUGCCGCGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGGUG-CCCAC--- ....(((((.(((((((((...--.)))))((((((.....((((((((....))))))))(((.....)))......)))))))))).))))).-.....--- ( -38.70, z-score = -0.68, R) >droSim1.chr3R 23812152 103 - 27517382 GUUGGCCAUGGCGAAUAUAGCCAUAUAUAUAGAGGCGAUGAGUGCCGCGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGGGG-CCCACUUU ((.(((((((((.......)))))........((((((((.((((((((....)))))))).))))))))(((((........))))).....))-)).))... ( -45.30, z-score = -2.48, R) >droSec1.super_22 732227 101 - 1066717 GUUGGCCAUGGCGAAUUUAGCC--AUAUAUAGAGGCGAUGAGUGCCGCGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGGGG-CCCACUUU ((.(((((((((.......)))--))......((((((((.((((((((....)))))))).))))))))(((((........))))).....))-)).))... ( -45.30, z-score = -2.55, R) >droYak2.chr3R 6456252 102 + 28832112 GUUGGCCAUGUAGAAUGGAGCC--AUAUAUACAGGCGAUGAGUGCCGUGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGUGGCCCCACUUU ...((((((((((((.((.(((--.......(((((((((.((((((((....)))))))).)))))))))..)))...)).))))))...))))))....... ( -40.20, z-score = -1.06, R) >droEre2.scaffold_4820 6550413 101 + 10470090 GUUGGCCAUGGUGAAUGGAGCC--AUACAGACAGGCGAUGAGUACCGCGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGGGG-CCCACUUU ((.((((((.(..((.((.((.--.(((...(((((((((.((.(((((....))))).)).)))))))))...))).)))).))..).))..))-)).))... ( -39.90, z-score = -0.91, R) >droGri2.scaffold_14830 4562415 90 - 6267026 --UAGUUGUG-UGUGUGUGGC---ACGGCAGCGGAUGU--AGUUUUAU-AGGGCGUGCCACGCAUUGUUCGUUGA-GCGCCUUUUAGUGGUG----CACACUUU --.....(((-((((((((((---(((.(....((...--....))..-..).))))))))))))..........-(((((.......))))----)))))... ( -34.90, z-score = -2.26, R) >consensus GUUGGCCAUGGCGAAUGUAGCC__AUAUAUACAGGCGAUGAGUGCCGCGGGGGCGUGGCACGCAUUGUCUGCAGGUAAGCCUUUUUGCGGUGGGG_CCCACUUU ...(((..((.....))..)))..........((((((((.((((((((....)))))))).))))))))(((((........)))))(((((....))))).. (-27.10 = -27.08 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:31 2011