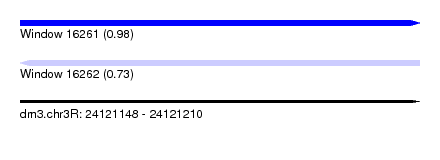
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,121,148 – 24,121,210 |
| Length | 62 |
| Max. P | 0.983352 |

| Location | 24,121,148 – 24,121,210 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 67.28 |
| Shannon entropy | 0.65694 |
| G+C content | 0.44427 |
| Mean single sequence MFE | -17.71 |
| Consensus MFE | -7.12 |
| Energy contribution | -7.42 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.983352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24121148 62 + 27905053 UUCAAGCCGCCAUCGUUGCCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUG---------- .....(((((((....))((((((((((..((((........))))))))))))))))))).---------- ( -20.00, z-score = -2.46, R) >droSim1.chr3R 23805064 62 + 27517382 UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUG---------- .....(((((((....))))((((((((..((((........)))))))))))).)))....---------- ( -20.20, z-score = -2.38, R) >droSec1.super_22 725282 62 + 1066717 UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUG---------- .....(((((((....))))((((((((..((((........)))))))))))).)))....---------- ( -20.20, z-score = -2.38, R) >droYak2.chr3R 6448886 62 - 28832112 UUUAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUG---------- .....(((((((....))))((((((((..((((........)))))))))))).)))....---------- ( -20.20, z-score = -2.27, R) >droEre2.scaffold_4820 6543219 62 - 10470090 UUCCAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGCUG---------- .....(((((((....))))((((((((..((((........)))))))))))).)))....---------- ( -20.20, z-score = -2.36, R) >droVir3.scaffold_13047 3789179 52 - 19223366 -UGCAAGCGGCCCCGUUUGUGUUAUUUAUGGGCGCUCACAUAUGCAUAAAUAA------------------- -.(((((((....))))))).(((((((((.(..........).)))))))))------------------- ( -12.60, z-score = -0.06, R) >droWil1.scaffold_181108 1368854 50 + 4707319 ----------CAACGUUUCUGUUAUUUAUUUGCGCUCACAUGUGCAUAAAUAACAUGUAU------------ ----------.........(((((((((..(((((......)))))))))))))).....------------ ( -13.10, z-score = -3.12, R) >dp4.chr2 22827207 65 - 30794189 -------CGCCACAGUCUGUGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGUGGACUGAGAGGGGGAG -------(.((.(((((..(((((((((..((((........)))))))))))))..)))))....)).).. ( -23.40, z-score = -3.88, R) >droPer1.super_0 10021897 65 - 11822988 -------CGCCACAGUCUGUGCUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGUGGACUGAGAGGGGGAG -------(.((.(((((..((.((((((..((((........)))))))))).))..)))))....)).).. ( -19.20, z-score = -1.99, R) >anoGam1.chr3R 6560650 62 - 53272125 AAUAGCGUGUCAUUUAUUUUUUGGUUCAUUUCCGGCACCACAUUCAAACACCACAAACGGUG---------- ....(.(((((......................)))))).........((((......))))---------- ( -8.05, z-score = -0.08, R) >consensus UUCAAGCCGCCAUCGUUGGCGUUAUUUAUUUGCGCUCACAUAUGCAUAAAUAACGGGCGGUG__________ ...................(((((((((..((((........)))))))))))))................. ( -7.12 = -7.42 + 0.30)

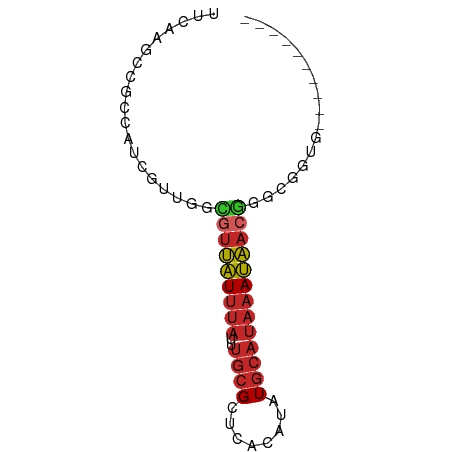
| Location | 24,121,148 – 24,121,210 |
|---|---|
| Length | 62 |
| Sequences | 10 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 67.28 |
| Shannon entropy | 0.65694 |
| G+C content | 0.44427 |
| Mean single sequence MFE | -15.49 |
| Consensus MFE | -5.91 |
| Energy contribution | -6.17 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24121148 62 - 27905053 ----------CACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGGCAACGAUGGCGGCUUGAA ----------..(((((.(((((((((.....(((....))).)))))))))(....)...)))))...... ( -18.40, z-score = -1.49, R) >droSim1.chr3R 23805064 62 - 27517382 ----------CACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUGAA ----------....(((.(((((((((.....(((....))).)))))))))((((....)))))))..... ( -17.80, z-score = -1.63, R) >droSec1.super_22 725282 62 - 1066717 ----------CACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUGAA ----------....(((.(((((((((.....(((....))).)))))))))((((....)))))))..... ( -17.80, z-score = -1.63, R) >droYak2.chr3R 6448886 62 + 28832112 ----------CACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUUAAA ----------....(((.(((((((((.....(((....))).)))))))))((((....)))))))..... ( -17.80, z-score = -1.81, R) >droEre2.scaffold_4820 6543219 62 + 10470090 ----------CAGCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACGCCAACGAUGGCGGCUGGAA ----------((((....(((((((((.....(((....))).)))))))))((((....)))).))))... ( -19.10, z-score = -1.34, R) >droVir3.scaffold_13047 3789179 52 + 19223366 -------------------UUAUUUAUGCAUAUGUGAGCGCCCAUAAAUAACACAAACGGGGCCGCUUGCA- -------------------.............((..(((((((................))).))))..))- ( -14.29, z-score = -1.33, R) >droWil1.scaffold_181108 1368854 50 - 4707319 ------------AUACAUGUUAUUUAUGCACAUGUGAGCGCAAAUAAAUAACAGAAACGUUG---------- ------------.....((((((((((((.(......).)))..))))))))).........---------- ( -9.50, z-score = -1.12, R) >dp4.chr2 22827207 65 + 30794189 CUCCCCCUCUCAGUCCACGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACACAGACUGUGGCG------- .....((...(((((...(((((((((.....(((....))).)))))))))...))))).))..------- ( -15.10, z-score = -2.36, R) >droPer1.super_0 10021897 65 + 11822988 CUCCCCCUCUCAGUCCACGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAGCACAGACUGUGGCG------- .....((...(((((...(((((((((.....(((....))).)))))))))...))))).))..------- ( -15.10, z-score = -1.77, R) >anoGam1.chr3R 6560650 62 + 53272125 ----------CACCGUUUGUGGUGUUUGAAUGUGGUGCCGGAAAUGAACCAAAAAAUAAAUGACACGCUAUU ----------...(((((.(((..((.......))..))).))))).......................... ( -10.00, z-score = 0.46, R) >consensus __________CACCGCCCGUUAUUUAUGCAUAUGUGAGCGCAAAUAAAUAACACAAACGAUGGCGGCUUGAA .................((((((((((.....(((....))).))))))))))................... ( -5.91 = -6.17 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:31 2011