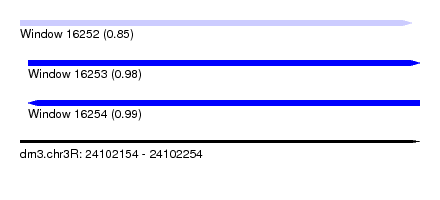
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,102,154 – 24,102,254 |
| Length | 100 |
| Max. P | 0.988646 |

| Location | 24,102,154 – 24,102,252 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 77.23 |
| Shannon entropy | 0.43628 |
| G+C content | 0.39209 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -7.09 |
| Energy contribution | -6.91 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
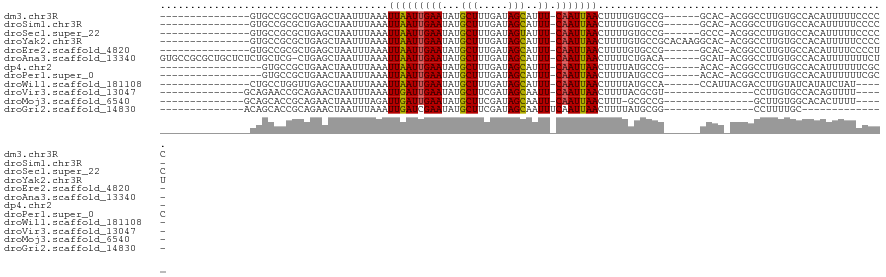
>dm3.chr3R 24102154 98 + 27905053 ---------------GUGCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUGUGCCG------GCAC-ACGGCCUUGUGCCACAUUUUUCCCCC ---------------(((((((((.(((..........(((((((((.(((((.....)))))))-))))))))))..))).))------))))-..(((.....)))............. ( -30.00, z-score = -4.08, R) >droSim1.chr3R 23786112 97 + 27517382 ---------------GUGCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUGUGCCG------GCAC-ACGGCCUUGUGCCACAUUUUUCCCC- ---------------(((((((((.(((..........(((((((((.(((((.....)))))))-))))))))))..))).))------))))-..(((.....)))............- ( -30.00, z-score = -4.04, R) >droSec1.super_22 706342 98 + 1066717 ---------------GUGCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGUAUUU-CAAUUAACUUUUGUGCCG------GCCC-ACGGCCUUGUGCCACAUUUUUCCCCC ---------------(((..((((.(((..........(((((((((.(((((.....)))))))-))))))).......((((------....-.))))))))))))))........... ( -24.10, z-score = -2.60, R) >droYak2.chr3R 6429668 104 - 28832112 ---------------GUGCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUGUGCCGCACAAGGCAC-ACGGCCUUGUGCCACAUUUUUCCCCU ---------------(((..((((.(((..........(((((((((.(((((.....)))))))-))))))))))..)))).(((((((((..-...))))))))))))........... ( -34.40, z-score = -4.64, R) >droEre2.scaffold_4820 6522545 97 - 10470090 ---------------GUGCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUGUGCCG------GCAC-ACGGCCUUGUGCCACAUUUUCCCCU- ---------------(((((((((.(((..........(((((((((.(((((.....)))))))-))))))))))..))).))------))))-..(((.....)))............- ( -30.00, z-score = -4.01, R) >droAna3.scaffold_13340 22230914 111 + 23697760 GUGCCGCGCUGCUCUCUGCUCG-CUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUCUGACA------GCAU-ACGGCCUUGUGCCACAUUUUUUUCU- ..((((.((((.((...((((.-..)))).........(((((((((.(((((.....)))))))-))))))).......))))------))..-.))))....................- ( -27.40, z-score = -2.32, R) >dp4.chr2 22808234 95 - 30794189 -----------------GUGCCGCUGAACUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUAUGCCG------ACAC-ACGGCCUUGUGCCACAUUUUUUCGC- -----------------(((.(((..............(((((((((.(((((.....)))))))-))))))).......((((------....-.))))...))).)))..........- ( -23.30, z-score = -3.48, R) >droPer1.super_0 10003083 96 - 11822988 -----------------GUGCCGCUGAACUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUAUGCCG------ACAC-ACGGCCUUGUGCCACAUUUUUUCGCC -----------------(((.(((..............(((((((((.(((((.....)))))))-))))))).......((((------....-.))))...))).)))........... ( -23.30, z-score = -3.58, R) >droWil1.scaffold_181108 1346825 94 + 4707319 ---------------CUGCCUGGUUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU-CAAUUAACUUUUAUGCCA------CCAUUACGACCUUGUAUCAUAUCUAU----- ---------------.(((..(((((............(((((((((.(((((.....)))))))-))))))).....(((...------.)))..)))))..)))..........----- ( -18.30, z-score = -2.45, R) >droVir3.scaffold_13047 3771571 86 - 19223366 --------------GCAGAACCGCAGAACUAAUUUAAAUUGAUUGAAUAUGCUUCGAUAGCAAUU-CAAUUAACUUUUACGCGU---------------CCUUGUGCCACAGUUUU----- --------------((......))((((((........((((((((((.((((.....)))))))-)))))))......((((.---------------...))))....))))))----- ( -17.60, z-score = -1.95, R) >droMoj3.scaffold_6540 15207428 85 - 34148556 --------------GCAGCACCGCAGAACUAAUUUAGAUUGAUUGAAUAUGCUUCGAUAGCAAUU-CAAUUAACUUU-GCGCCG---------------GCUUGUGGCACACUUUU----- --------------(((((..((((((.(((...))).((((((((((.((((.....)))))))-))))))).)))-)))...---------------)).)))...........----- ( -21.60, z-score = -1.80, R) >droGri2.scaffold_14830 4540734 78 + 6267026 --------------ACAGCACCGCAGAACUAAUUUAAAUUGAUCGAAUAUGCUUCGAUAGCAAUUUCAAUUAACUUUUAUGCGG---------------CCUUUUGC-------------- --------------...((((((((....(((((.((((((((((((.....))))))..)))))).))))).......)))))---------------.....)))-------------- ( -17.00, z-score = -2.14, R) >consensus _______________CUGCCCCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUU_CAAUUAACUUUUGUGCCG______GCAC_ACGGCCUUGUGCCACAUUUUUCCCC_ ......................................(((((((....((((.....))))....)))))))................................................ ( -7.09 = -6.91 + -0.18)

| Location | 24,102,156 – 24,102,254 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Shannon entropy | 0.40645 |
| G+C content | 0.38904 |
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.19 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24102156 98 + 27905053 --------------GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUGUGCC------GGCAC-ACGGCCUUGUGCCACAUUUUUCCCCCAC --------------...((((.(((..........(((((((((.(((((.....)))))))))))))))))..)))).------(((((-(......))))))............... ( -25.80, z-score = -2.94, R) >droSim1.chr3R 23786114 97 + 27517382 --------------GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUGUGCC------GGCAC-ACGGCCUUGUGCCACAUUUUUCCCCAC- --------------...((((.(((..........(((((((((.(((((.....)))))))))))))))))..)))).------(((((-(......))))))..............- ( -25.80, z-score = -2.89, R) >droSec1.super_22 706344 98 + 1066717 --------------GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGUAUUUCAAUUAACUUUUGUGCC------GGCCC-ACGGCCUUGUGCCACAUUUUUCCCCCAC --------------((((.((((.((......((.(((((((((.(((((.....)))))))))))))).))....)))------)))..-.))))....................... ( -22.90, z-score = -2.34, R) >droYak2.chr3R 6429670 104 - 28832112 --------------GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUGUGCCGCACAAGGCAC-ACGGCCUUGUGCCACAUUUUUCCCCUAU --------------...((.....)).........(((((((((.(((((.....))))))))))))))....((((..(((((((((..-...)))))))))))))............ ( -32.50, z-score = -4.44, R) >droEre2.scaffold_4820 6522547 97 - 10470090 --------------GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUGUGCC------GGCAC-ACGGCCUUGUGCCACAUUUU-CCCCUAU --------------...((((.(((..........(((((((((.(((((.....)))))))))))))))))..)))).------(((((-(......)))))).......-....... ( -25.80, z-score = -2.87, R) >droAna3.scaffold_13340 22230916 111 + 23697760 GCCGCGCUGCUCUCUGCUCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUCUGAC------AGCAU-ACGGCCUUGUGCCACAUUUUUUUCUGU- ((((.((((.((...((((...)))).........(((((((((.(((((.....)))))))))))))).......)))------)))..-.))))......................- ( -27.40, z-score = -2.32, R) >dp4.chr2 22808236 93 - 30794189 ----------------GCCGCUGAACUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUAUGCC------GACAC-ACGGCCUUGUGCCACAUUUUUUCGC--- ----------------(.(((..............(((((((((.(((((.....)))))))))))))).......(((------(....-.))))...))).)............--- ( -19.00, z-score = -2.43, R) >droPer1.super_0 10003085 96 - 11822988 ----------------GCCGCUGAACUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUAUGCC------GACAC-ACGGCCUUGUGCCACAUUUUUUCGCCAU ----------------...((.(((..(((.....(((((((((.(((((.....)))))))))))))).......(((------(....-.))))..........)))..)))))... ( -19.30, z-score = -2.38, R) >droWil1.scaffold_181108 1346827 92 + 4707319 --------------GCCUGGUUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUAUGCC------ACCAUUACGACCUUGUAUCAUAUCUAU------- --------------((..(((((............(((((((((.(((((.....)))))))))))))).....(((..------..)))..)))))..))...........------- ( -17.30, z-score = -2.27, R) >droVir3.scaffold_13047 3771573 84 - 19223366 -------------AGAACCGCAGAACUAAUUUAAAUUGAUUGAAUAUGCUUCGAUAGCAAUUCAAUUAACUUUU--------------AC-GCGUCCUUGUGCCACAGUUUU------- -------------........((((((........((((((((((.((((.....)))))))))))))).....--------------.(-(((....))))....))))))------- ( -16.40, z-score = -2.00, R) >droMoj3.scaffold_6540 15207430 83 - 34148556 -------------AGCACCGCAGAACUAAUUUAGAUUGAUUGAAUAUGCUUCGAUAGCAAUUCAAUUAACUUU---------------GC-GCCGGCUUGUGGCACACUUUU------- -------------......(((((.(((...))).((((((((((.((((.....)))))))))))))).)))---------------))-((((.....))))........------- ( -20.70, z-score = -1.93, R) >consensus ______________GCCGCGCUGAGCUAAUUUAAAUUAAUUGAAUAUGCUUUGAUAGCAUUUCAAUUAACUUUUGUGCC______GGCAC_ACGGCCUUGUGCCACAUUUUUCCCC_A_ ...................................(((((((((.(((((.....))))))))))))))........................(((.....)))............... (-12.92 = -13.19 + 0.27)

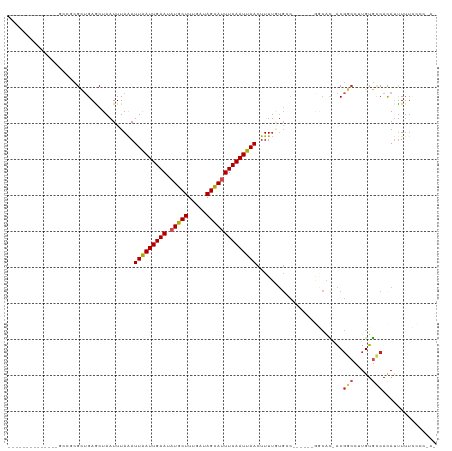
| Location | 24,102,156 – 24,102,254 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Shannon entropy | 0.40645 |
| G+C content | 0.38904 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
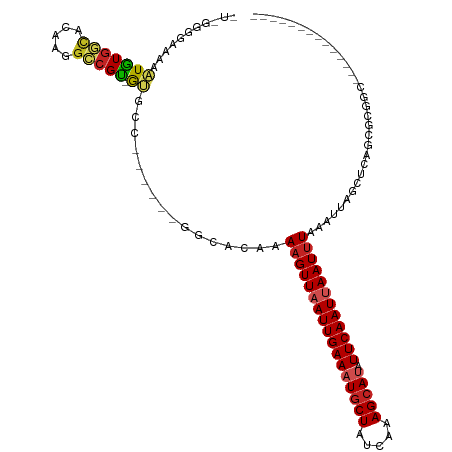
>dm3.chr3R 24102156 98 - 27905053 GUGGGGGAAAAAUGUGGCACAAGGCCGU-GUGCC------GGCACAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC-------------- (((.(((.....(((((((((......)-)))))------.)))...(((((((((((((((((.....))))).)))))))))))).......)))..)))...-------------- ( -30.20, z-score = -2.66, R) >droSim1.chr3R 23786114 97 - 27517382 -GUGGGGAAAAAUGUGGCACAAGGCCGU-GUGCC------GGCACAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC-------------- -(((.............)))...(((((-((...------(((....(((((((((((((((((.....))))).))))))))))))......)))..)))))))-------------- ( -28.92, z-score = -2.34, R) >droSec1.super_22 706344 98 - 1066717 GUGGGGGAAAAAUGUGGCACAAGGCCGU-GGGCC------GGCACAAAAGUUAAUUGAAAUACUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC-------------- (((.(((.....((((.(....((((..-.))))------.))))).((((((((((((((.((.....)).)).)))))))))))).......)))..)))...-------------- ( -22.90, z-score = -0.79, R) >droYak2.chr3R 6429670 104 + 28832112 AUAGGGGAAAAAUGUGGCACAAGGCCGU-GUGCCUUGUGCGGCACAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC-------------- ...(((......(((.(((((((((...-..))))))))).)))...(((((((((((((((((.....))))).)))))))))))).......)))........-------------- ( -37.60, z-score = -4.32, R) >droEre2.scaffold_4820 6522547 97 + 10470090 AUAGGGG-AAAAUGUGGCACAAGGCCGU-GUGCC------GGCACAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC-------------- .......-...............(((((-((...------(((....(((((((((((((((((.....))))).))))))))))))......)))..)))))))-------------- ( -28.30, z-score = -2.39, R) >droAna3.scaffold_13340 22230916 111 - 23697760 -ACAGAAAAAAAUGUGGCACAAGGCCGU-AUGCU------GUCAGAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGAGCAGAGAGCAGCGCGGC -......................(((((-.((((------.((....(((((((((((((((((.....))))).))))))))))))......((((...)))).)).))))..))))) ( -35.90, z-score = -4.38, R) >dp4.chr2 22808236 93 + 30794189 ---GCGAAAAAAUGUGGCACAAGGCCGU-GUGUC------GGCAUAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGUUCAGCGGC---------------- ---(((((...((((((((((......)-)))))------.))))..(((((((((((((((((.....))))).)))))))))))).......))).))...---------------- ( -26.60, z-score = -3.66, R) >droPer1.super_0 10003085 96 + 11822988 AUGGCGAAAAAAUGUGGCACAAGGCCGU-GUGUC------GGCAUAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGUUCAGCGGC---------------- (((.(((....(..((((.....)))).-.).))------).)))..(((((((((((((((((.....))))).))))))))))))................---------------- ( -28.40, z-score = -3.76, R) >droWil1.scaffold_181108 1346827 92 - 4707319 -------AUAGAUAUGAUACAAGGUCGUAAUGGU------GGCAUAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAACCAGGC-------------- -------.....((((((.....)))))).((((------(((.((((((((((((((((((((.....))))).))))))))))))...))))))..))))...-------------- ( -23.80, z-score = -3.93, R) >droVir3.scaffold_13047 3771573 84 + 19223366 -------AAAACUGUGGCACAAGGACGC-GU--------------AAAAGUUAAUUGAAUUGCUAUCGAAGCAUAUUCAAUCAAUUUAAAUUAGUUCUGCGGUUCU------------- -------..((((((......(((((..-..--------------..(((((.(((((((((((.....)))).))))))).)))))......)))))))))))..------------- ( -18.60, z-score = -1.86, R) >droMoj3.scaffold_6540 15207430 83 + 34148556 -------AAAAGUGUGCCACAAGCCGGC-GC---------------AAAGUUAAUUGAAUUGCUAUCGAAGCAUAUUCAAUCAAUCUAAAUUAGUUCUGCGGUGCU------------- -------.....((((((.......)))-))---------------)..(((.(((((((((((.....)))).))))))).))).....................------------- ( -17.40, z-score = -0.38, R) >consensus _U_GGGGAAAAAUGUGGCACAAGGCCGU_GUGCC______GGCACAAAAGUUAAUUGAAAUGCUAUCAAAGCAUAUUCAAUUAAUUUAAAUUAGCUCAGCGCGGC______________ ..............((((.....))))....................(((((((((((((((((.....))))).))))))))))))................................ (-14.88 = -15.58 + 0.70)

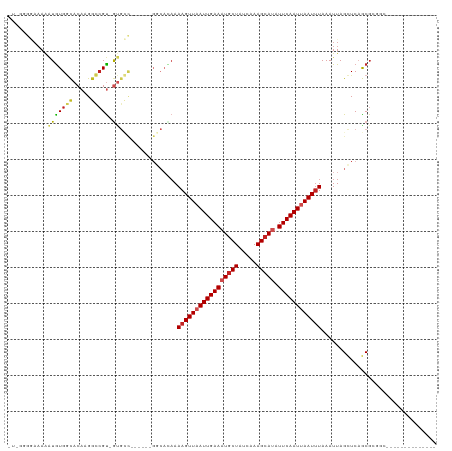
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:24 2011