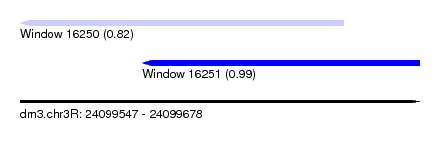
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,099,547 – 24,099,678 |
| Length | 131 |
| Max. P | 0.993955 |

| Location | 24,099,547 – 24,099,653 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Shannon entropy | 0.40364 |
| G+C content | 0.44552 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -19.61 |
| Energy contribution | -21.14 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.816418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24099547 106 - 27905053 UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAAGAGUCAGC---------GAGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGCUGCAUCGUGGUAACU .........(((((((((((..(((.((((((.....)))))).))).)))))))))---------))(((((((.(..(((..((.......))..)))..)....))))))). ( -33.90, z-score = -1.75, R) >droSim1.chr3R 23783529 106 - 27517382 UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAGGAGUCAGC---------GAGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGCUGCACCGUGGUAACU .........(((((((((((..(((.((((((.....)))))).))).)))))))))---------))(((((((.(..(((..((.......))..)))..)....))))))). ( -33.80, z-score = -1.36, R) >droSec1.super_22 703977 106 - 1066717 UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAGGAGUCAGC---------GAGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGCUGCACCGUGGUAACU .........(((((((((((..(((.((((((.....)))))).))).)))))))))---------))(((((((.(..(((..((.......))..)))..)....))))))). ( -33.80, z-score = -1.36, R) >droYak2.chr3R 6426893 106 + 28832112 UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAGGAGUCGGC---------GGGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGUUGCACCUUGGUAACU ..........((((((((((..(((.((((((.....)))))).))).)))))))))---------)((((((((((..((..(((..........)))..))..)))))))))) ( -35.90, z-score = -2.56, R) >droEre2.scaffold_4820 6519954 106 + 10470090 UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUGGCAGAUGGAGCCGGAGUCGGC---------GAGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGUUGCACCUUGGUAACU .........(((((((((((..(((.((((((.....)))))).))).)))))))))---------))(((((((((..((..(((..........)))..))..))))))))). ( -36.60, z-score = -2.39, R) >droAna3.scaffold_13340 22228396 114 - 23697760 UAAGAAUUUUUGUUGGCUUAACGCUGCAUUUGGA-AGCAGAUGGAGCGGGAUGUGGCAGAUACAGAGAGUUAGCGAGUGGCUUUGGUGGAUUUUGUCAGGUGCACCUAGGUUACU .........((((((((((..((((.((((((..-..)))))).))))...((((.....))))..))))))))))(((((((.((((.(((......))).)))).))))))). ( -40.90, z-score = -3.98, R) >droWil1.scaffold_181089 8280906 108 - 12369635 GAAAACCUUUAGUGGCCAUUUCGAAAGAUUCGCAUUGCAUAU-UAGUUAGUUCUUUCA------GUCGAUCUUUGAGUUGUGUUUUUGUUAUUUGUCACAAGCUUCUAGCUAACU ...........(((((....((....))...(((..(((((.-.....((.((.....------...)).))......)))))...))).....))))).(((.....))).... ( -16.60, z-score = 0.90, R) >consensus UAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAGGAGUCAGC_________GAGUUAUCAAGUGGCUUUUGUGGAUUUUGUCAGCUGCACCUUGGUAACU ...........(((((((((..(((.((((((.....)))))).))).)))))))))..........((((((((....((..(((..........)))..))....)))))))) (-19.61 = -21.14 + 1.54)

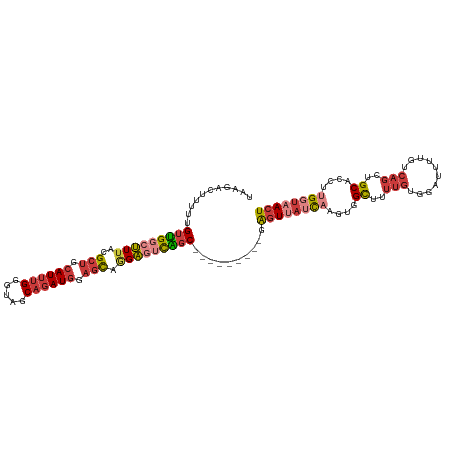
| Location | 24,099,587 – 24,099,678 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.55949 |
| G+C content | 0.45879 |
| Mean single sequence MFE | -28.99 |
| Consensus MFE | -18.07 |
| Energy contribution | -17.76 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.993955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 24099587 91 - 27905053 ---------CUGGGGAAUGCCUGGGGU-UUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAA--GAGUCAGCGAGUUAUCA--------- ---------(..((.....))..)(((-(((.....)))))).((((((((((((..(((.((((((.....)))))).))).)--)))))))))))......--------- ( -30.20, z-score = -2.82, R) >droSim1.chr3R 23783569 99 - 27517382 CUGUCUGCUCUGGGGAAUGCCUGGGG--UUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAG--GAGUCAGCGAGUUAUCA--------- ..((((((((..((.....))..)))--)........))))..((((((((((((..(((.((((((.....)))))).))).)--)))))))))))......--------- ( -34.80, z-score = -2.43, R) >droSec1.super_22 704017 81 - 1066717 --------------------CUGGGGUGUUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAG--GAGUCAGCGAGUUAUCA--------- --------------------....((((((((....)))))..((((((((((((..(((.((((((.....)))))).))).)--)))))))))))..))).--------- ( -25.00, z-score = -2.61, R) >droYak2.chr3R 6426933 100 + 28832112 UUGUCUGCUCUGGGAAAUGCCUGGGGU-UUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAG--GAGUCGGCGGGUUAUCA--------- ..((((((((..((.....))..))))-.........))))..((((((((((((..(((.((((((.....)))))).))).)--)))))))))))......--------- ( -32.90, z-score = -2.18, R) >droEre2.scaffold_4820 6519994 99 + 10470090 UUGUCUGCUCGGGGGAAAGCCUGGGG--UUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUGGCAGAUGGAGCCG--GAGUCGGCGAGUUAUCA--------- (((((.((((.(...((((((.((((--((((....)))))))).....))))))..(((.((((((.....)))))).)))).--)))).))))).......--------- ( -33.10, z-score = -1.57, R) >droAna3.scaffold_13340 22228436 104 - 23697760 ---UUGGCGCCAGGGGAUGCCAGGGG--UUUUUAAUAAGAAUUUUUGUUGGCUUAACGCUGCAUUUGGA-AGCAGAUGGAGCGG--GAUGUGGCAGAUACAGAGAGUUAGCG ---..((((((...)).))))(((((--((((.....)))))))))((((((((..((((.((((((..-..)))))).)))).--..((((.....))))..)))))))). ( -32.10, z-score = -2.59, R) >droWil1.scaffold_181089 8280946 93 - 12369635 ---------CAAGGUGAGGCCUGUUGU-UUAUUAAGAAAACCUUUAGUGGCCAUUUCGAAAGAUUCGCAUUGCAUAUUAGUUAGUUCUUUCAGUCGAUCUUUG--------- ---------.(((((((((((....((-((.......)))).......)))).....((((((...((...((......))..))))))))..)).)))))..--------- ( -14.80, z-score = 0.73, R) >consensus ___U__GC_CUGGGGAAUGCCUGGGGU_UUUUUAAUAAGACUUUUUGUUGGCUUUACGCUGCAUUUGGGUAGCAGAUGGAGCAG__GAGUCAGCGAGUUAUCA_________ ......................................((...(((((((((((...(((.((((((.....)))))).)))....)))))))))))...)).......... (-18.07 = -17.76 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:22 2011