| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,095,383 – 24,095,482 |
| Length | 99 |
| Max. P | 0.909601 |

| Location | 24,095,383 – 24,095,482 |
|---|---|
| Length | 99 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 64.76 |
| Shannon entropy | 0.72349 |
| G+C content | 0.50349 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -10.28 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
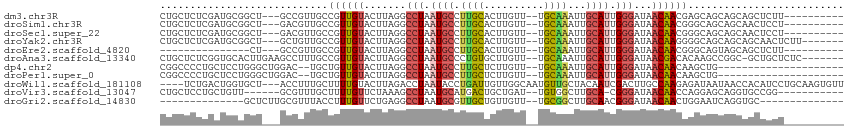
>dm3.chr3R 24095383 99 - 27905053 CUGCUCUCGAUGCGGCU---GCCGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU--UGCAAAUUGCAUUGGGAUAACAACGAGCAGCAGCAGCUCUU---------- ..((.......))((((---((.(((((((((((...((..((((((((.(((((......--)))))...)))))))).)))))))).))))).))))))...---------- ( -41.40, z-score = -3.45, R) >droSim1.chr3R 23779221 99 - 27517382 CUGCUCUCGAUGCGGCU---GACGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU--UGCAAAUUGCAUUGGGAUAACAACGGGCAGCAGCAACUCCU---------- (((((((((..(((((.---......)))))..........((((((((.(((((......--)))))...)))))))).......)))).)))))........---------- ( -34.30, z-score = -1.83, R) >droSec1.super_22 699867 99 - 1066717 CUGCUCUCGAUGCGGCU---GACGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU--UGCAAAUUGCAUUGGGAUAACAACGGGCAGCAGCAACUCCU---------- (((((((((..(((((.---......)))))..........((((((((.(((((......--)))))...)))))))).......)))).)))))........---------- ( -34.30, z-score = -1.83, R) >droYak2.chr3R 6421526 102 + 28832112 CUGCUCUCGAUGCGGCU---GCUGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU--UGCAAAUUGCAUUGGGAUAACAAGGGGCAGCAGCAGCAACUCUU------- .(((.......)))(((---(((((((((.((....(((..((((((((.(((((......--)))))...)))))))).)))..)).)))))))))))).......------- ( -43.20, z-score = -3.14, R) >droEre2.scaffold_4820 6515666 84 + 10470090 ---------------CU---GCCGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU--UGCAAAUUGCAUUGGGAUAACAACGGGCAGUAGCAGCUCUU---------- ---------------((---((..((((((((((...((..((((((((.(((((......--)))))...)))))))).))))))).)))))..)))).....---------- ( -33.60, z-score = -3.12, R) >droAna3.scaffold_13340 22224164 104 - 23697760 CUGCUCUCGGUGCACUUGAAGCCUUUGCCGUUGUACUUAGGCCUAAUGCCCUGUGCUUGUU--UGCAAAUUGCAUUGGGAUAACGACACAAGCCGGC-GCUGCUCUC------- .......((((((.((((..((....)).(((((...((..((((((((....(((.....--.)))....)))))))).))))))).))))...))-)))).....------- ( -30.50, z-score = -0.31, R) >dp4.chr2 22801094 89 + 30794189 CGGCCCCUGCUCCUGGGCUGGAC--UGCUGUUGUACUUAGGCCUAAUGCCUUGCUCUUGUU--UGCAAAUUGCAUUGGGAUAACAACAAGCUG--------------------- ((((((........))))))...--.((((((((...((..((((((((.((((.......--.))))...)))))))).))))))).)))..--------------------- ( -31.60, z-score = -2.32, R) >droPer1.super_0 9995906 89 + 11822988 CGGCCCCUGCUCCUGGGCUGGAC--UGCUGUUGUACUUAGGCCUAAUGCCUUGCUCUUGUU--UGCAAAUUGCAUUGGGAUAACAACAAGCUG--------------------- ((((((........))))))...--.((((((((...((..((((((((.((((.......--.))))...)))))))).))))))).)))..--------------------- ( -31.60, z-score = -2.32, R) >droWil1.scaffold_181108 1329421 107 - 4707319 ----UCUGACUGGUGCU---ACCUUUGCUUUUGUACUUAGACCUAAUACCUGAUUGUUGGCAAUGUUGCUACAAUCGACUUGCCAAGAGAUAAUAACCACAUCCUGCAAGUGUU ----((((...(((((.---............))))))))).........(((((((.((((....)))))))))))((((((..((.(((.........)))))))))))... ( -21.32, z-score = -0.56, R) >droVir3.scaffold_13047 3764042 94 + 19223366 CUGCUCCUGCUGUU------GCGUUUGCUUUUGUUCUAAAGCCUAAUGCAUGACUGCUGAU--UGUGGCUUGCA-CGGGAUAACAACCAGGAGCAGGUGCCGG----------- (((((((((.((((------(.....(((((......)))))((..((((.(.(..(....--.)..)).))))-..)).)))))..))))))))).......----------- ( -32.20, z-score = -1.31, R) >droGri2.scaffold_14830 4533980 84 - 6267026 --------------GCUCUUGCGUUUACCUUUGUUCUGAGGCCUAAUGCGUUGCUGUUGUU--UGCGGCUUGCAACGGGAUAACAACUGGAAUCAGGUGC-------------- --------------.(((..(((((..((((......))))...)))))(((((.((((..--..))))..))))))))....((.(((....))).)).-------------- ( -22.50, z-score = -0.01, R) >consensus CUGCUCUCGAUGCGGCU___GCCGUUGCCGUUGUACUUAGGCCUAAUGCCUUGCACUUGUU__UGCAAAUUGCAUUGGGAUAACAACGGGCAGCAGCAGCUCCU__________ ............................((((((.......(((.((((.((((..........))))...)))).)))...)))))).......................... (-10.28 = -10.36 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:20 2011