
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,094,341 – 24,094,441 |
| Length | 100 |
| Max. P | 0.955489 |

| Location | 24,094,341 – 24,094,441 |
|---|---|
| Length | 100 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 61.72 |
| Shannon entropy | 0.81038 |
| G+C content | 0.43551 |
| Mean single sequence MFE | -27.49 |
| Consensus MFE | -7.67 |
| Energy contribution | -7.61 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
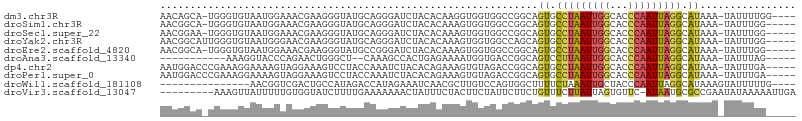
>dm3.chr3R 24094341 100 - 27905053 AACAGCA-UGGGUGUAAUGGAAACGAAGGGUAUGCAGGGAUCUACACAAGGUGGUGGCCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUUGG---- ....((.-(((((((..((....))..((((((((..((..((((.....))))...)).))).))))).....))))))).....)).....-........---- ( -30.30, z-score = -1.29, R) >droSim1.chr3R 23778185 99 - 27517382 AACGGCA-UGGGUGUAAUGGAAACGAAGGGUAUGCAGGGAUCUACACAAAGUGGUGGCCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGG----- ..((((.-(((((.(........(....)........).)))))(((......)))))))...((((((((((((...))))))))))))...-.......----- ( -30.49, z-score = -1.42, R) >droSec1.super_22 698842 99 - 1066717 AACGGAA-UGGGUGUAAUGGAAACGAAGGGUAUGCAGGGAUCUACACAAAGUGGUGGCCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGG----- .......-(((((((..((....))..((((((((..((..((((.....))))...)).))).))))).....)))))))............-.......----- ( -29.30, z-score = -1.54, R) >droYak2.chr3R 6420531 100 + 28832112 AACGGCAUUGGGUGUAAUGGGAACGAAGGGUAUGCAGGGAUCUACACAAAGUGGUGGCCAGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGG----- ....(((((((((((..((....))..((((((((..((..((((.....))))...)).))).))))).....)))))))))...)).....-.......----- ( -29.80, z-score = -1.13, R) >droEre2.scaffold_4820 6514648 99 + 10470090 AACGGCA-UGGGUGUAAUGGAAACGAAGGGUAUGCCGGGAUCUACACAAAGUGGUGGCCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGG----- ....((.-(((((((..((....))..(((((((((((...((((.....))))...)))))).))))).....))))))).....)).....-.......----- ( -35.90, z-score = -2.61, R) >droAna3.scaffold_13340 22223129 87 - 23697760 -----------AAAGGUACCCAGAACUGGGCU--CAAAGCCACUGAGAAAAUGGUGACCGGCAGUCCUUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUAG----- -----------...(((..(((...((.(((.--....)))....))....)))..)))....((.(((((((((...))))))))).))...-.......----- ( -19.00, z-score = 0.18, R) >dp4.chr2 22799970 100 + 30794189 AAUGGACCCGAAAGGAAAAGUAGGAAAGUCCUACCAAAUCUACACAGAAAGUGUAGACCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGA----- ..(((..((....))....(((((.....)))))....(((((((.....))))))))))...((((((((((((...))))))))))))...-.......----- ( -37.80, z-score = -6.24, R) >droPer1.super_0 9994786 100 + 11822988 AAUGGACCCGAAAGGAAAAGUAGGAAAGUCCUACCAAAUCUACACAGAAAGUGUAGACCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA-UAUUUGA----- ..(((..((....))....(((((.....)))))....(((((((.....))))))))))...((((((((((((...))))))))))))...-.......----- ( -37.80, z-score = -6.24, R) >droWil1.scaffold_181108 1328563 87 - 4707319 ---------------AACGGUCGACUGCCAUAGACCAUAGAAAUCAACGCUUGUCCAGUGGCUUUUCUAAAUUGCUACCCAAUUAGGCAUAAAGUAUUUUUG---- ---------------...((((..........))))............(((((....(((((...........))))).....)))))..............---- ( -15.90, z-score = -0.40, R) >droVir3.scaffold_13047 3763245 96 + 19223366 ---------AAAGUUAUUUUUGUGGUAUCUUUUGAAAAAAACUAUUUCUACUUCUAUUCUUCUGUUUCUUAUUAGUGUUC-AUAAUGCGCCGAAUAUAAAAAUUGA ---------....((((.((((..((((....((((....((((.....((............)).......)))).)))-)..))))..)))).))))....... ( -8.60, z-score = 0.48, R) >consensus AACGGCA_UGGGUGUAAUGGAAACGAAGGGUAUGCAAGGAUCUACACAAAGUGGUGGCCGGCAGUGCCUAAUUGGCACCCAAUUAGGCAUAAA_UAUUUGG_____ ...............................................................((.(((((((((...))))))))).))................ ( -7.67 = -7.61 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:19 2011