| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,077,098 – 24,077,194 |
| Length | 96 |
| Max. P | 0.775816 |

| Location | 24,077,098 – 24,077,194 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.86 |
| Shannon entropy | 0.54790 |
| G+C content | 0.46769 |
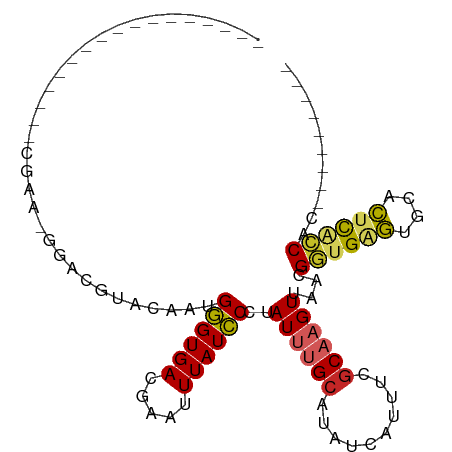
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -12.76 |
| Energy contribution | -12.28 |
| Covariance contribution | -0.48 |
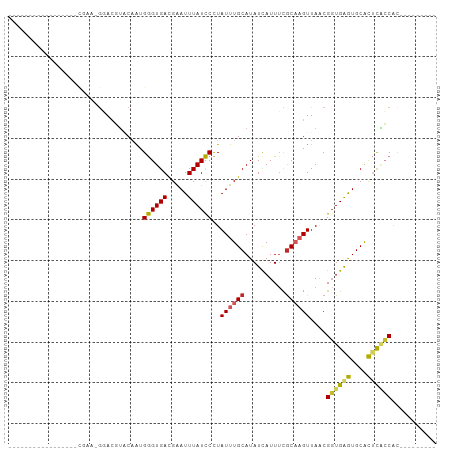
| Combinations/Pair | 1.33 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.775816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
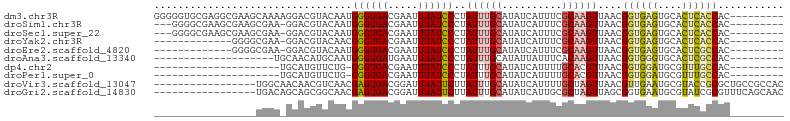
>dm3.chr3R 24077098 96 + 27905053 GGGGGUGCGAGGCGAAGCAAAAGGACGUACAAUGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCACCAC--------- .....(((((((.((.((((((((..(((.(.(.(....).).).))).))).)))))...)).))))))).......((((((....))))))..--------- ( -27.10, z-score = -1.23, R) >droSim1.chr3R 23763918 92 + 27517382 ---GGGGCGAAGCGAAGCGAA-GGACGUACAAUGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCACCAC--------- ---...((((((.((.((...-.).)(((.((((((.((........)))))))))))...)).))))))........((((((....))))))..--------- ( -26.50, z-score = -1.57, R) >droSec1.super_22 684321 92 + 1066717 ---GGGGCGAAGCGAAGCGAA-GGACGUACAAUGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCACCAC--------- ---...((((((.((.((...-.).)(((.((((((.((........)))))))))))...)).))))))........((((((....))))))..--------- ( -26.50, z-score = -1.57, R) >droYak2.chr3R 6405481 82 - 28832112 -------------GGGGCGAA-GGACGUACAACGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCACCAC--------- -------------...(((((-(((..(.(((.((((((.....))))))....))).)..)).))))))........((((((....))))))..--------- ( -23.40, z-score = -1.77, R) >droEre2.scaffold_4820 6498815 82 - 10470090 -------------GGGGCGAA-GGACGUACAAUGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCGCCAC--------- -------------...(((((-(((.(((.((((((.((........)))))))))))...)).))))))........((((((....))))))..--------- ( -24.10, z-score = -1.58, R) >droAna3.scaffold_13340 22211163 76 + 23697760 --------------------UGCAACAUGCAAUGGGUGAUGAAUUUAUCCCUAUUUGCAUAUUAUUUCACAAGUUAACGGUGGGUGCACUCGCCAC--------- --------------------......((((((((((.((((....)))))))).))))))..................((((((....))))))..--------- ( -21.60, z-score = -1.79, R) >dp4.chr2 22785989 74 - 30794189 ---------------------UGCAUGUUCUG-GGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUUGCACGUUAACGGUGGAUGCGUUUGCCAC--------- ---------------------((((.((...(-((((((.....))))))).)).))))...................((..((....))..))..--------- ( -17.30, z-score = -0.12, R) >droPer1.super_0 9980702 74 - 11822988 ---------------------UGCAUGUUCUG-GGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUUGCACGUUAACGGUGGAUGCGUUUGCCAC--------- ---------------------((((.((...(-((((((.....))))))).)).))))...................((..((....))..))..--------- ( -17.30, z-score = -0.12, R) >droVir3.scaffold_13047 3750657 88 - 19223366 -----------------UGGCAACAACGUCAACGAGUGACGGAUUUAUUCUUAUUUGCAUAUCAUUUUGCUAGUUAACGUUGAAUGCGUACCGCGCUGCCGCCAC -----------------.((((.((((((.(((((((((.....))))))......(((........)))..))).))))))...(((.....)))))))..... ( -25.00, z-score = -1.67, R) >droGri2.scaffold_14830 4520541 88 + 6267026 -----------------UGACAGCAGCGGCAACGAGUGACGGAUUUAUUCUUAUUUGCAUAUCAUUGCGCUAGUUAGCGGUGAAUGCGUAUCGCGUUUCAGCAAC -----------------(((.(((.((((..((((((((.....))))))......((((.(((((((........))))))))))))).))))))))))..... ( -22.90, z-score = 0.10, R) >consensus _________________CGAA_GGACGUACAAUGGGUGACGAAUUUAUCCCUAUUUGCAUAUCAUUUCGCAAGUUAACGGUGAGUGCACUCACCAC_________ .................................((((((.....))))))..((((((..........))))))....((((((....))))))........... (-12.76 = -12.28 + -0.48)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:17 2011