| Sequence ID | dm3.chr2L |
|---|---|
| Location | 8,867,760 – 8,867,858 |
| Length | 98 |
| Max. P | 0.813335 |

| Location | 8,867,760 – 8,867,858 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 66.64 |
| Shannon entropy | 0.62154 |
| G+C content | 0.45184 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -12.79 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
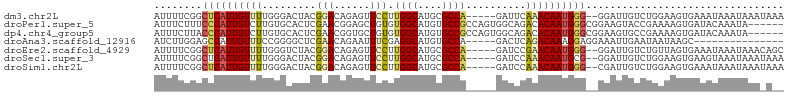
>dm3.chr2L 8867760 98 + 23011544 AUUUUCGGCUCAUUGUUUUGGGACUACGGACAGAGUUCCUUGGCAUGCGCCA-----GAUUCAAACAAUGGG--GGAUUGUCUGGAAGUGAAAUAAAUAAAUAAA ((((((((((((((((((.((((((.(.....)))))))(((((....))))-----)....))))))))))--.......))))))))................ ( -26.41, z-score = -1.66, R) >droPer1.super_5 1646308 99 - 6813705 AUUUCUUUCCCAUUGUCUUGUGCACUCGAACGGAGCUGUGUGGCAUGUGCCGCCAGUGGCAGACACAAUGGGCGGAAGUACCGAAAAGUGAUACAAAUA------ ((((((..((((((((..(.(((.(((.....)))(((.(((((....))))))))..))).).)))))))).))))))....................------ ( -31.60, z-score = -1.14, R) >dp4.chr4_group5 1675201 99 - 2436548 AUUUCUUACCCAUUGUCUUGUGCACUCGAACGGUGCUGUGUGGCAUGUGCCGCCAGUGGCAGACACAAUGGGCGGAAGUGCCGAAAAGUGAUACAAAUA------ ((((((..((((((((..(.((((((.....)))((((.(((((....))))))))).))).).)))))))).))))))....................------ ( -31.30, z-score = -0.47, R) >droAna3.scaffold_12916 10532477 85 + 16180835 AUCUUGGAGCCAUUGUUUCCGGGGCUCGAACAGAAUUUCGAGGCAUGUGCCA-----GACUCAGACAAAGGAGGAAAUUGAAUAAUAAGC--------------- ..(((((((((.(((....))))))))......((((((..(((....))).-----..(((........)))))))))......)))).--------------- ( -19.80, z-score = -0.18, R) >droEre2.scaffold_4929 9460988 98 + 26641161 AUUUUCGGCUCAUUGUUUUGGGUCUACGGACAGAGUUCCUUGGCAUGCGCCA-----GAUCCGAACAAUGGG--GGAUUGUCUGUUAGUGAAAUAAAUAAACAGC ........((((((((((.((((((..(((......)))..(((....))))-----)))))))))))))))--.......(((((.((.......)).))))). ( -29.20, z-score = -1.73, R) >droSec1.super_3 4341878 98 + 7220098 AUUUUCGGCUCAUUGUUUUGGGACUACGGACAGAGUUCCUUGGCAUGCGCCA-----GAUCCAAACAAUGCG--GGAUUGUCUGGAAGUGAAGUAAAUAAAUAAA ..(((((((.(((.(((..((((((.(.....)))))))..)))))).))).-----..((((.(((((...--..))))).))))...))))............ ( -27.90, z-score = -1.57, R) >droSim1.chr2L 8646741 98 + 22036055 AUUUUCGGCUCAUUGUUUUGGGACUACGGACAGAGUUCCUUGGCAUGCGCCA-----GAUCCAAACAAUGGG--CGAUUGUCUGGAAGUGAAAUAAAUAAAUAAA ..(((((((.(((.(((..((((((.(.....)))))))..)))))).))).-----..((((.(((((...--..))))).))))...))))............ ( -28.20, z-score = -1.95, R) >consensus AUUUUCGGCUCAUUGUUUUGGGACUACGGACAGAGUUCCUUGGCAUGCGCCA_____GAUCCAAACAAUGGG__GGAUUGUCUGAAAGUGAAAUAAAUAAA_A__ ........((((((((((.........(((......)))..(((....)))...........))))))))))................................. (-12.79 = -13.04 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:27:09 2011