| Sequence ID | dm3.chr3R |
|---|---|
| Location | 24,046,388 – 24,046,479 |
| Length | 91 |
| Max. P | 0.983675 |

| Location | 24,046,388 – 24,046,479 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Shannon entropy | 0.41090 |
| G+C content | 0.37175 |
| Mean single sequence MFE | -17.93 |
| Consensus MFE | -13.50 |
| Energy contribution | -12.91 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
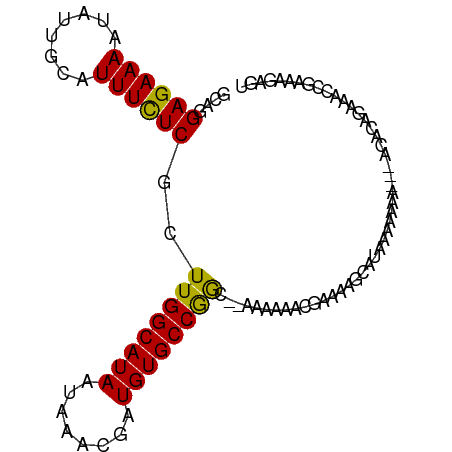
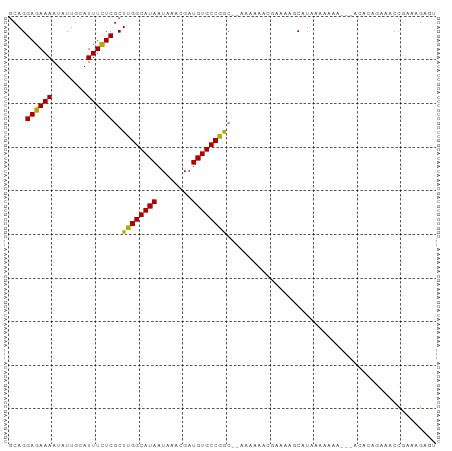
>dm3.chr3R 24046388 91 - 27905053 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC--AAAAAACGAAAAGCAUAAAAA-----ACACAGAAACCGAAAGAGU ((..((((((........))))))(((.((((((........)))))))))--............)).......-----........(((....).)) ( -19.60, z-score = -2.37, R) >droSim1.chr3R 23736047 91 - 27517382 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC--AAAAAACGAAAAGCAUAAAAA-----ACACAGAAACCGAAAGAGU ((..((((((........))))))(((.((((((........)))))))))--............)).......-----........(((....).)) ( -19.60, z-score = -2.37, R) >droSec1.super_22 658763 91 - 1066717 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC--AAAAAACGAAAAGCAUAAAAA-----ACACAGAAACCGAAAGAGU ((..((((((........))))))(((.((((((........)))))))))--............)).......-----........(((....).)) ( -19.60, z-score = -2.37, R) >droYak2.chr3R 6378730 97 + 28832112 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC-AAAAAAAUGAAAAGCAUAAAAAAAAACACACAGAAACCGAAAGAGU ((..((((((........))))))(((.((((((........)))))))))-.............))....................(((....).)) ( -19.60, z-score = -2.35, R) >droEre2.scaffold_4820 6472697 90 + 10470090 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC--AAAAAACGAAAAGCAU-GAAA-----ACACAGAAACCGAAAGAGU ((..((((((........))))))(((.((((((........)))))))))--............))..-....-----........(((....).)) ( -19.60, z-score = -2.02, R) >dp4.chr2 22759149 92 + 30794189 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGAC--AAAAAAAAAAUACCAUCCAGAAAG--GAAGAAAAAUUGGACGU-- ....((((((........))))))(.((((((((........)))))))))--............((((((.....)--))........)))....-- ( -17.00, z-score = -1.27, R) >droPer1.super_0 9953567 94 + 11822988 GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGACAAAAAAAAAAAAUACCAUCCAGAAAG--GAAGAAAAAUUGGACGU-- ....((((((........))))))(.((((((((........)))))))))..............((((((.....)--))........)))....-- ( -17.00, z-score = -1.29, R) >droVir3.scaffold_13047 3724649 85 + 19223366 ACAGGAGAAAAUAUUGCAUUUUUCGCUUGGCAUAAUAAACGAUGUGCCAAAAAAA-CCACAAAACCC-UCGAAAAAA--AAAAACAAAC--------- ...((.(((((........)))))..((((((((........)))))))).....-)).........-.........--..........--------- ( -14.00, z-score = -2.43, R) >droMoj3.scaffold_6540 15155033 81 + 34148556 ACAGGAGAAAAUAUUGCAUUUUUCGCUUGGCAUAAUAAACGAUGUGCCAAAAAAAGCCACAAAAACC-UCGAAAAAG--CAACG-------------- .............((((.(((((((.((((((((........)))))))).................-.))))))))--)))..-------------- ( -15.41, z-score = -1.84, R) >consensus GCAGGAGAAAAUAUUGCAUUUCUCGCUUGGCAUAAUAAACGAUGUGCCGGC__AAAAAACGAAAAGCAUAAAAAAA___ACACAGAAACCGAAAGAGU ....((((((........))))))..((((((((........))))))))................................................ (-13.50 = -12.91 + -0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:15 2011