| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,987,350 – 23,987,447 |
| Length | 97 |
| Max. P | 0.985325 |

| Location | 23,987,350 – 23,987,447 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.87 |
| Shannon entropy | 0.39687 |
| G+C content | 0.35844 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -12.41 |
| Energy contribution | -12.41 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
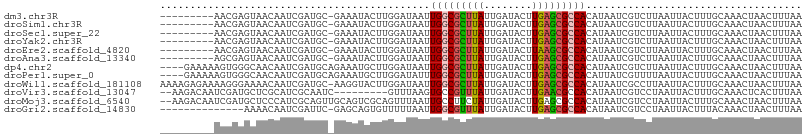
>dm3.chr3R 23987350 97 - 27905053 ---------AACGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------..(((.......))).(((-(((.((...(((..(((((((((((........)))))))))....))..)))....)).))))))............ ( -23.10, z-score = -3.39, R) >droSim1.chr3R 23681823 97 - 27517382 ---------AACGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------..(((.......))).(((-(((.((...(((..(((((((((((........)))))))))....))..)))....)).))))))............ ( -23.10, z-score = -3.39, R) >droSec1.super_22 606012 97 - 1066717 ---------AACGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------..(((.......))).(((-(((.((...(((..(((((((((((........)))))))))....))..)))....)).))))))............ ( -23.10, z-score = -3.39, R) >droYak2.chr3R 6325151 97 + 28832112 ---------AACGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------..(((.......))).(((-(((.((...(((..(((((((((((........)))))))))....))..)))....)).))))))............ ( -23.10, z-score = -3.39, R) >droEre2.scaffold_4820 6417003 97 + 10470090 ---------AACGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUAAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------..(((.......))).(((-(((.((...(((..(((((((((((........)))))))))....))..)))....)).))))))............ ( -23.00, z-score = -3.58, R) >droAna3.scaffold_13340 22130068 97 - 23697760 ---------AGCGAGUAACAAUCGAUGC-GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ---------.((((((((....((((.(-((.....)))......(((((((((........)))))))))....))))......)))).))))............. ( -22.80, z-score = -2.51, R) >dp4.chr2 22702547 103 + 30794189 ----GAAAAAGUGGGCAACAAUCGAUGCAGAAAUGCUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA ----.....(((..((((....(((.(((....))))))(((.(((((((((((........))))))))).)).)))............))))..)))........ ( -28.40, z-score = -3.57, R) >droPer1.super_0 9897405 103 + 11822988 ----GAAAAAGUGGGCAACAAUCGAUGCAGAAAUGCUUGGAUAUUUGGCGCUUAUUGAUACUUGAGCGCCACAUUAUCGUUUUAAUUACUUUGCAAACUAACUUUAA ----.....(((..((((....(((.(((....))))))((((..(((((((((........)))))))))...))))............))))..)))........ ( -30.20, z-score = -4.11, R) >droWil1.scaffold_181108 1226233 106 - 4707319 AAAAGAGAAAAGGGAAAACAAUCGAUGC-AAGGUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGCCUUAAUUACUUUGCAAACUAACUUUAA ........((((.((......))..(((-((((((...((.....(((((((((........)))))))))........)).....)))))))))......)))).. ( -26.52, z-score = -3.20, R) >droVir3.scaffold_13047 3670302 96 + 19223366 --AAGACAAUCGAUGCUCGCAUCGCAAUC---------GUUUAAGUGCCGUUUAUUGAUACUUGAACGCCACAUAAUCGUCCUAAUUACUUUGCAAACUCACUUUAA --..(((...(((((....)))))....(---------(((((((((.((.....)).))))))))))..........))).......................... ( -18.60, z-score = -1.77, R) >droMoj3.scaffold_6540 15091806 105 + 34148556 --AAGACAAUCGAUGCUCCCAUCGCAGUUGCAGUCGCAGUUUAAUUGCCUUCUAUUGAUACUUGAGCGCCACAUAAUCGUCCUAAUUACUUUGCAAACUAACUUUAA --..(((....((((....))))((....)).)))((((..(((((((.(((...........))).)).((......))...)))))..))))............. ( -14.50, z-score = 0.62, R) >droGri2.scaffold_14830 4437098 92 - 6267026 --------------AAAACAAUCGAUUC-GAGCAGUGUUUUUAAUUGGCGUUUAUUGAUACUUGAGCGCCACAUAAUCGUCCUAAUUACUUUACAAACUAACUUUAA --------------((((((.(((...)-))....))))))....(((((((((........))))))))).................................... ( -15.50, z-score = -1.23, R) >consensus _________AACGAGUAACAAUCGAUGC_GAAAUACUUGGAUAAUUGGCGCUUAUUGAUACUUGAGCGCCACAUAAUCGUCUUAAUUACUUUGCAAACUAACUUUAA .............................................(((((((((........))))))))).................................... (-12.41 = -12.41 + -0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:10 2011