
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,954,396 – 23,954,499 |
| Length | 103 |
| Max. P | 0.651180 |

| Location | 23,954,396 – 23,954,499 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Shannon entropy | 0.45639 |
| G+C content | 0.47119 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.47 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
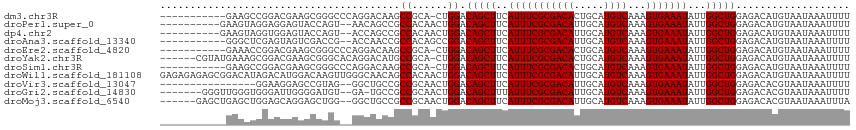
>dm3.chr3R 23954396 103 + 27905053 -----------GAAGCCGGACGAAGCGGGCCCAGGACAAGCCGCA-CUGGACAGCUUCAUUUCGCGACACUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU -----------..((((((..(((((..(.((((..(.....)..-)))).).)))))(((((((((((.....))))...))))))).))))))(....).............. ( -30.60, z-score = -1.56, R) >droPer1.super_0 9860085 103 - 11822988 ----------GAAGUAGGAGGAGUACCAGU--AACAGCCGCCACAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU ----------(((((.((((..((.(((((--.............)))))))..))))))))).....((((((((((..(((.(.....).)))...))))))))))....... ( -27.12, z-score = -1.81, R) >dp4.chr2 22666400 103 - 30794189 ----------GAAGUAGGUGGAGUACCAGU--ACCAGCCGCCACAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU ----------(((((.(((((.(((....)--))...))))).(....)....)))))..........((((((((((..(((.(.....).)))...))))))))))....... ( -27.40, z-score = -1.34, R) >droAna3.scaffold_13340 22097272 102 + 23697760 -----------GGGCUCGAGUAGUCGACCG--ACCAACCGCCACAGCCGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU -----------.((((((.((.(((....)--))..))))....))))...(((((..((((((((((((...)))))...)))))))...)))))................... ( -24.70, z-score = -0.12, R) >droEre2.scaffold_4820 6383851 103 - 10470090 -----------GAAACCGGACGAAGCGGGCCCAGGACAAGCCGCA-CUGGACAGCUUCAUUUCGCGACACUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU -----------....(((((.(((((..(.((((..(.....)..-)))).).)))))..)))).)....((((((((..(((.(.....).)))...))))))))......... ( -27.10, z-score = -0.88, R) >droYak2.chr3R 6290974 108 - 28832112 ------CGUAUGAAAGCGGACGAAGCGGGCACAGGACAUGCCGCA-CUGGACAGCUUCAUUUCGCGACACUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU ------.........(((((.(((((..(..(((..(.....)..-)))..).)))))..))))).....((((((((..(((.(.....).)))...))))))))......... ( -29.20, z-score = -1.11, R) >droSim1.chr3R 23654509 103 + 27517382 -----------GAAGCCGGACGAAGCGGGCCCAGGACAAGCCGCA-CUGGACAGCUUCAUUUCGCGACACUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU -----------..((((((..(((((..(.((((..(.....)..-)))).).)))))(((((((((((.....))))...))))))).))))))(....).............. ( -30.60, z-score = -1.56, R) >droWil1.scaffold_181108 1189748 115 + 4707319 GAGAGAGAGCGGACAUAGACAUGGACAAGUUGGGCAACAGCCACAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU ((((..(((((..(((....)))..).((((((((....))).))))).....))))..)))).....((((((((((..(((.(.....).)))...))))))))))....... ( -35.10, z-score = -3.86, R) >droVir3.scaffold_13047 3634651 97 - 19223366 ----------------GGAAGGAGCCGUAG--GGCUGCCGCCGCAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACACGUAAUAAAUUUU ----------------((..(.((((....--)))).)..))...((((..(((((..((((((((((((...)))))...)))))))...)))))....)).)).......... ( -26.70, z-score = -0.50, R) >droGri2.scaffold_14830 4400796 105 + 6267026 -------GGGUUGGGUGGGAUUGGGGAUGU--GA-UGCCGCCGCAACUGGACAGCUUUAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACACGUAAUAAAUUUU -------.((((((((((.(((........--))-).))))).)))))...(((((.(((((((((((((...)))))...))))))))..)))))................... ( -34.10, z-score = -2.66, R) >droMoj3.scaffold_6540 15052890 107 - 34148556 ------GAGCUGAGCUGGAGCAGGAGCUGG--GGCUGCCGCCGCAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACACGUAAUAAAUUUA ------.((((((((((...(((....((.--(((....))).)).)))..)))))).((((((((((((...)))))...)))))))...))))(....).............. ( -30.80, z-score = 0.04, R) >consensus ___________GAGAUCGAACGGAGCGGGC__AGCAGCCGCCGCAACUGGACAGCUUCAUUUCGCGACAUUGCAUGUCAAAGUGAAAUAUUGGCUGGAGACAUGUAAUAAAUUUU ........................................(((....))).(((((..(((((((((((.....))))...)))))))...)))))................... (-16.55 = -16.47 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:06 2011