| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,952,545 – 23,952,636 |
| Length | 91 |
| Max. P | 0.996852 |

| Location | 23,952,545 – 23,952,636 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.88 |
| Shannon entropy | 0.52936 |
| G+C content | 0.51206 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -19.73 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.996852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
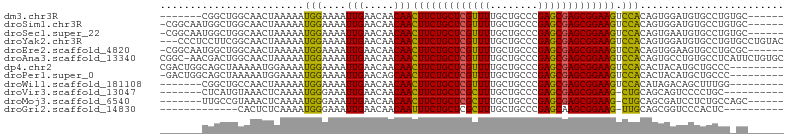
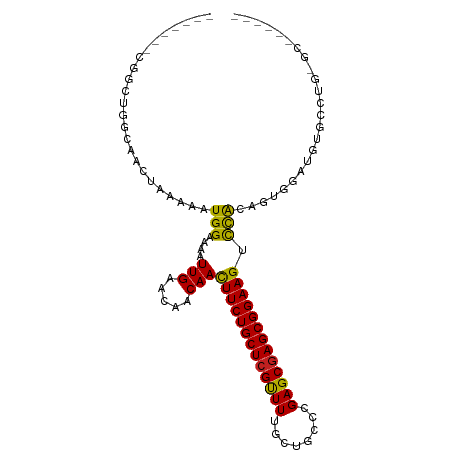
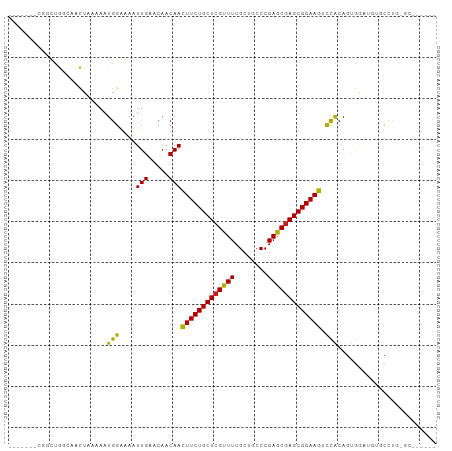
>dm3.chr3R 23952545 91 + 27905053 -------CGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGAUGUGCCUGUGC------ -------..((.((((.(((....((((...(((.....)))(((((((((((((........)))))))))))))))))...)))...)))).))..------ ( -31.00, z-score = -2.13, R) >droSim1.chr3R 23652593 97 + 27517382 -CGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGAUGUGCCUGUGC------ -..(((..(((..(((.(((....((((...(((.....)))(((((((((((((........)))))))))))))))))...))).))))))..)))------ ( -32.40, z-score = -1.74, R) >droSec1.super_22 571411 97 + 1066717 -CGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGAAUGUGCCUGUGC------ -.((((.(.((((....(((....)))..............((((((((((((((........))))))))))))))...)))).)...)))).....------ ( -31.20, z-score = -1.72, R) >droYak2.chr3R 6288946 101 - 28832112 ---CCCUCCUUCGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGAUGUGCCUGUGCCUGUAC ---.........((((.(((....((((...(((.....)))(((((((((((((........)))))))))))))))))...)))...))))........... ( -29.80, z-score = -1.87, R) >droEre2.scaffold_4820 6381896 97 - 10470090 -CGGCAAUGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGAAGUGCCUGCGC------ -........((.((((.(((....((((...(((.....)))(((((((((((((........)))))))))))))))))...)))...)))).))..------ ( -33.00, z-score = -1.61, R) >droAna3.scaffold_13340 22095459 103 + 23697760 CGGC-AACGACUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGCCUGUGCCUCAUUCUGUGC .(((-(......((((.((.....((((...(((.....)))(((((((((((((........))))))))))))))))).))))))..))))........... ( -32.70, z-score = -1.88, R) >dp4.chr2 22664671 95 - 30794189 CGACUGGCAGCUAAAAAUGGAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACACUACAUGCUGCCC--------- .....((((((.....(((.....((((...(((.....)))(((((((((((((........))))))))))))))))).....))))))))).--------- ( -33.40, z-score = -4.20, R) >droPer1.super_0 9858313 94 - 11822988 -GACUGGCAGCUAAAAAUGGAAAAUGGAAAAUUGAACAGCAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACACUACAUGCUGCCC--------- -....((((((.....(((.....((((...(((.....)))(((((((((((((........))))))))))))))))).....))))))))).--------- ( -33.60, z-score = -3.73, R) >droWil1.scaffold_181108 1188588 88 + 4707319 -------CGGCUGCCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAUAGACAGCUUUGG--------- -------.((((((((........)))..............((((((((((((((........)))))))))))))).........)))))....--------- ( -27.50, z-score = -2.53, R) >droVir3.scaffold_13047 3633144 86 - 19223366 -------CUCAUGUAAACUCAAAAUGGGAAAUUGAACAACAACUUCUGCUCGCUUUGCUGCCCGAGCGAGCGGAAG-CUGCAGCAGUCCCCUGC---------- -------(((((...........)))))...........((.(((((((((((((........)))))))))))))-.))..((((....))))---------- ( -28.30, z-score = -2.25, R) >droMoj3.scaffold_6540 15051386 90 - 34148556 -------UUGCCGUAAACUCAAAAUGGGAAAUUGAACAACAACUUCUGCUCGCUUUGCUGCCCGAGCGAGCGGAAG-CUGCAGCGAUCCUCUGCCAGC------ -------((((.(((..(((.....)))..............(((((((((((((........)))))))))))))-.))).))))............------ ( -27.90, z-score = -1.24, R) >droGri2.scaffold_14830 4399235 80 + 6267026 -------------CACUCUCAAAAUGGGAAAUUGAACAACAAUUUCUGCUCGCUUUGCUGCCCGAGCAAGCGGAAG-UUGCAGCGGUCCCACUC---------- -------------...........(((((...((.....(((((((((((.((((........)))).))))))))-)))...)).)))))...---------- ( -25.10, z-score = -1.92, R) >consensus _______CGGCUGGCAACUAAAAAUGGAAAAUUGAACAACAACUUCUGCUCGUUUUGCUGCCCGAGCGAGCGGAAGUCCACAGUGGAUGUGCCUG_GC______ ........................(((....(((.....)))(((((((((((((........))))))))))))).)))........................ (-19.73 = -19.10 + -0.63)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:05 2011