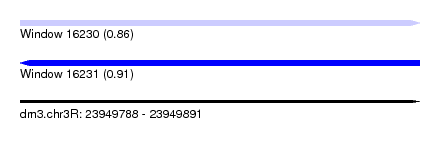
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 23,949,788 – 23,949,891 |
| Length | 103 |
| Max. P | 0.912047 |

| Location | 23,949,788 – 23,949,891 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.56194 |
| G+C content | 0.44596 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -13.19 |
| Energy contribution | -14.28 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
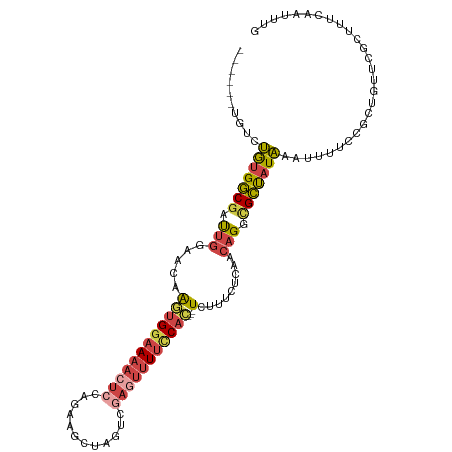
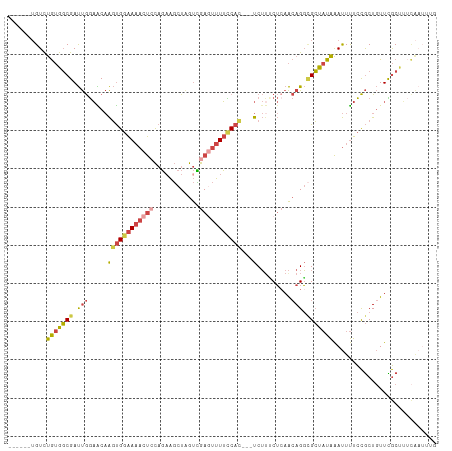
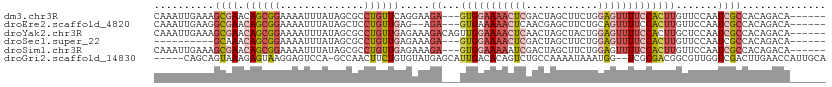
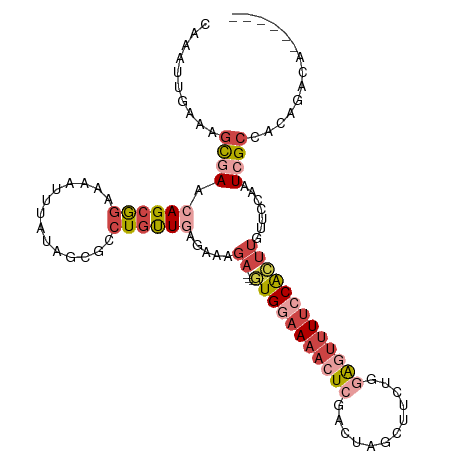
>dm3.chr3R 23949788 103 + 27905053 ------UGUCUGUGGCGAUUGGAACAAGUGGAAAACUCCAGAAGCUAGUCGAGUUUUCCAC---UCUUCCUGAACAGGCGCUAUAAAUUUUCCGCUGUUCGCUUUCAAUUUG ------....((.(((....((((..((((((((((((..((......)))))))))))))---).)))).((((((.((..(......)..)))))))))))..))..... ( -31.60, z-score = -2.59, R) >droEre2.scaffold_4820 6379104 101 - 10470090 ------UGUCUGUGGCGAUUGGAACAAGUGGAAAACUGCAGAAGCUCGUUGAGUUUUUCAC---UCU--CUCAACAGGAGCUAUAAAUUUUCCGCUGUUCGCCUUCAAUUUG ------....((.(((((.....(((.((((((((.......(((((((((((........---...--))))))..))))).....))))))))))))))))..))..... ( -29.40, z-score = -1.89, R) >droYak2.chr3R 6286308 106 - 28832112 ------UGUCUGUGGCGAUUGGAGCAAGUGGAAAACUCCAGUAGCUAGUUGAGUUUUCCAACUGUCUUUCUCAACAGGCGCUAUAAAUUUUCCGCUGUUCGCUUUCAAUUUG ------.((.....))((..((((((.((((((((....(((.(((.((((((................)))))).)))))).....))))))))))))).)..))...... ( -28.79, z-score = -1.54, R) >droSec1.super_22 568639 93 + 1066717 ------UGUCUGUGGCGAUUGGAACAAGUGGAAAACUCCAGAAGCUAGUCGAGUUUUCCAC---UCUUUCUCAACAGGCGCUAUAAAUUUUCCGCUGUUUGC---------- ------....(((((((.((((((..((((((((((((..((......)))))))))))))---)..))).)))....)))))))........((.....))---------- ( -28.70, z-score = -2.38, R) >droSim1.chr3R 23649784 103 + 27517382 ------UGUCUGUGGCGAUUGGAACAAGUGGAAAACUCCAGAAGCUAGUCGAUUUUUCCAC---UCUUUCUCAACAGGCGCUAUAAAUUUUCCGCUGUUCGCUUUCAAUUUG ------....((.(((((((((((..(((((((((.((..((......)))).))))))))---)..))).)))..((((..(......)..))))..)))))..))..... ( -23.70, z-score = -0.76, R) >droGri2.scaffold_14830 4395063 104 + 6267026 UGCAAUGGUUCAAGUCGACCAACGCCGUCGCGA--CCAUUUAUUUUGGCAGACUGUGUCAAUGCUCAUACACAGAAGUUGGC-UGGACUCCUUACUCUUUACUGCUG----- .(((..(((...((((.((((((((((......--..........))))...((((((..........))))))..))))).-).))))....)))......)))..----- ( -23.79, z-score = 0.16, R) >consensus ______UGUCUGUGGCGAUUGGAACAAGUGGAAAACUCCAGAAGCUAGUCGAGUUUUCCAC___UCUUUCUCAACAGGCGCUAUAAAUUUUCCGCUGUUCGCUUUCAAUUUG ..........(((((((.(((......(((((((((((............))))))))))).............))).)))))))........................... (-13.19 = -14.28 + 1.09)

| Location | 23,949,788 – 23,949,891 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.76 |
| Shannon entropy | 0.56194 |
| G+C content | 0.44596 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -12.60 |
| Energy contribution | -14.52 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 23949788 103 - 27905053 CAAAUUGAAAGCGAACAGCGGAAAAUUUAUAGCGCCUGUUCAGGAAGA---GUGGAAAACUCGACUAGCUUCUGGAGUUUUCCACUUGUUCCAAUCGCCACAGACA------ ..........((((((((((............)).)))))).((((((---(((((((((((.(........).))))))))))))).))))....))........------ ( -33.90, z-score = -3.38, R) >droEre2.scaffold_4820 6379104 101 + 10470090 CAAAUUGAAGGCGAACAGCGGAAAAUUUAUAGCUCCUGUUGAG--AGA---GUGAAAAACUCAACGAGCUUCUGCAGUUUUCCACUUGUUCCAAUCGCCACAGACA------ .........(((((((((.((((((((...(((((..((((((--...---........))))))))))).....)))))))).).))).....))))).......------ ( -29.20, z-score = -2.47, R) >droYak2.chr3R 6286308 106 + 28832112 CAAAUUGAAAGCGAACAGCGGAAAAUUUAUAGCGCCUGUUGAGAAAGACAGUUGGAAAACUCAACUAGCUACUGGAGUUUUCCACUUGCUCCAAUCGCCACAGACA------ ..........((((..((((((((((((.((((....((((((.....(....).....))))))..))))...)))))))))....)))....))))........------ ( -25.50, z-score = -1.25, R) >droSec1.super_22 568639 93 - 1066717 ----------GCAAACAGCGGAAAAUUUAUAGCGCCUGUUGAGAAAGA---GUGGAAAACUCGACUAGCUUCUGGAGUUUUCCACUUGUUCCAAUCGCCACAGACA------ ----------((.....))............(((...((((.(((.((---(((((((((((.(........).))))))))))))).))))))))))........------ ( -28.00, z-score = -2.47, R) >droSim1.chr3R 23649784 103 - 27517382 CAAAUUGAAAGCGAACAGCGGAAAAUUUAUAGCGCCUGUUGAGAAAGA---GUGGAAAAAUCGACUAGCUUCUGGAGUUUUCCACUUGUUCCAAUCGCCACAGACA------ ..........((((.((((((..............)))))).(((.((---((((((((.((.(........).)).)))))))))).)))...))))........------ ( -25.14, z-score = -1.09, R) >droGri2.scaffold_14830 4395063 104 - 6267026 -----CAGCAGUAAAGAGUAAGGAGUCCA-GCCAACUUCUGUGUAUGAGCAUUGACACAGUCUGCCAAAAUAAAUGG--UCGCGACGGCGUUGGUCGACUUGAACCAUUGCA -----..(((((......((((..(.(((-(((......(((((..........)))))((((((((.......)))--.)).))))))..))).)..))))....))))). ( -26.90, z-score = -0.06, R) >consensus CAAAUUGAAAGCGAACAGCGGAAAAUUUAUAGCGCCUGUUGAGAAAGA___GUGGAAAACUCGACUAGCUUCUGGAGUUUUCCACUUGUUCCAAUCGCCACAGACA______ ..........((((.((((((..............))))))..........(((((((((((............))))))))))).........)))).............. (-12.60 = -14.52 + 1.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:50:04 2011